Genome-wide genotype and sequence-based reconstruction of the 140,000 year history of modern human ancestry
- PMID: 25116736
- PMCID: PMC4131216
- DOI: 10.1038/srep06055
Genome-wide genotype and sequence-based reconstruction of the 140,000 year history of modern human ancestry
Abstract
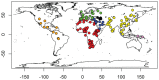
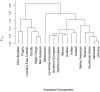
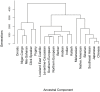
We investigated ancestry of 3,528 modern humans from 163 samples. We identified 19 ancestral components, with 94.4% of individuals showing mixed ancestry. After using whole genome sequences to correct for ascertainment biases in genome-wide genotype data, we dated the oldest divergence event to 140,000 years ago. We detected an Out-of-Africa migration 100,000-87,000 years ago, leading to peoples of the Americas, east and north Asia, and Oceania, followed by another migration 61,000-44,000 years ago, leading to peoples of the Caucasus, Europe, the Middle East, and south Asia. We dated eight divergence events to 33,000-20,000 years ago, coincident with the Last Glacial Maximum. We refined understanding of the ancestry of several ethno-linguistic groups, including African Americans, Ethiopians, the Kalash, Latin Americans, Mozabites, Pygmies, and Uygurs, as well as the CEU sample. Ubiquity of mixed ancestry emphasizes the importance of accounting for ancestry in history, forensics, and health.
Figures


References
-
- Li J. Z. et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science 319, 1100–1104 (2008). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

