Most human non-GCIMP glioblastoma subtypes evolve from a common proneural-like precursor glioma
- PMID: 25117714
- PMCID: PMC4143139
- DOI: 10.1016/j.ccr.2014.06.005
Most human non-GCIMP glioblastoma subtypes evolve from a common proneural-like precursor glioma
Abstract
To understand the relationships between the non-GCIMP glioblastoma (GBM) subgroups, we performed mathematical modeling to predict the temporal sequence of driver events during tumorigenesis. The most common order of evolutionary events is 1) chromosome (chr) 7 gain and chr10 loss, followed by 2) CDKN2A loss and/or TP53 mutation, and 3) alterations canonical for specific subtypes. We then developed a computational methodology to identify drivers of broad copy number changes, identifying PDGFA (chr7) and PTEN (chr10) as driving initial nondisjunction events. These predictions were validated using mouse modeling, showing that PDGFA is sufficient to induce proneural-like gliomas and that additional NF1 loss converts proneural to the mesenchymal subtype. Our findings suggest that most non-GCIMP mesenchymal GBMs arise as, and evolve from, a proneural-like precursor.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures

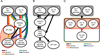


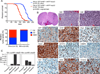


Comment in
-
RESICstance is futile-but not in glioblastoma.Cancer Cell. 2014 Aug 11;26(2):156-7. doi: 10.1016/j.ccr.2014.07.013. Cancer Cell. 2014. PMID: 25117706
References
-
- Baeza N, Weller M, Yonekawa Y, Kleihues P, Ohgaki H. PTEN methylation and expression in glioblastomas. Acta neuropathologica. 2003;106:479–485. - PubMed
-
- Beerenwinkel N, Sullivant S. Markov models for accumulating mutations. Biometrika. 2009;96:645–861.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

