Copy number variation in Thai population
- PMID: 25118596
- PMCID: PMC4131886
- DOI: 10.1371/journal.pone.0104355
Copy number variation in Thai population
Abstract
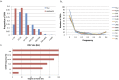
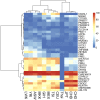
Copy number variation (CNV) is a major genetic polymorphism contributing to genetic diversity and human evolution. Clinical application of CNVs for diagnostic purposes largely depends on sufficient population CNV data for accurate interpretation. CNVs from general population in currently available databases help classify CNVs of uncertain clinical significance, and benign CNVs. Earlier studies of CNV distribution in several populations worldwide showed that a significant fraction of CNVs are population specific. In this study, we characterized and analyzed CNVs in 3,017 unrelated Thai individuals genotyped with the Illumina Human610, Illumina HumanOmniexpress, or Illumina HapMap550v3 platform. We employed hidden Markov model and circular binary segmentation methods to identify CNVs, extracted 23,458 CNVs consistently identified by both algorithms, and cataloged these high confident CNVs into our publicly available Thai CNV database. Analysis of CNVs in the Thai population identified a median of eight autosomal CNVs per individual. Most CNVs (96.73%) did not overlap with any known chromosomal imbalance syndromes documented in the DECIPHER database. When compared with CNVs in the 11 HapMap3 populations, CNVs found in the Thai population shared several characteristics with CNVs characterized in HapMap3. Common CNVs in Thais had similar frequencies to those in the HapMap3 populations, and all high frequency CNVs (>20%) found in Thai individuals could also be identified in HapMap3. The majorities of CNVs discovered in the Thai population, however, were of low frequency, or uniquely identified in Thais. When performing hierarchical clustering using CNV frequencies, the CNV data were clustered into Africans, Europeans, and Asians, in line with the clustering performed with single nucleotide polymorphism (SNP) data. As CNV data are specific to origin of population, our population-specific reference database will serve as a valuable addition to the existing resources for the investigation of clinical significance of CNVs in Thais and related ethnicities.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

