Polysialylated N-glycans identified in human serum through combined developments in sample preparation, separations, and electrospray ionization-mass spectrometry
- PMID: 25118826
- PMCID: PMC4151788
- DOI: 10.1021/ac501839b
Polysialylated N-glycans identified in human serum through combined developments in sample preparation, separations, and electrospray ionization-mass spectrometry
Abstract
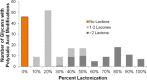
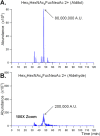
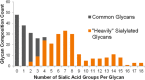
The N-glycan diversity of human serum glycoproteins, i.e., the human blood serum N-glycome, is both complex and constrained by the range of glycan structures potentially synthesizable by human glycosylation enzymes. The known glycome, however, has been further limited by methods of sample preparation, available analytical platforms, e.g., based upon electrospray ionization-mass spectrometry (ESI-MS), and software tools for data analysis. In this report several improvements have been implemented in sample preparation and analysis to extend ESI-MS glycan characterization and to include polysialylated N-glycans. Sample preparation improvements included acidified, microwave-accelerated, PNGase F N-glycan release to promote lactonization, and sodium borohydride reduction, that were both optimized to improve quantitative yields and conserve the number of glycoforms detected. Two-stage desalting (during solid phase extraction and on the analytical column) increased sensitivity by reducing analyte signal division between multiple reducing-end-forms or cation adducts. Online separations were improved by using extended length graphitized carbon columns and adding TFA as an acid modifier to a formic acid/reversed phase gradient, providing additional resolving power and significantly improved desorption of both large and heavily sialylated glycans. To improve MS sensitivity and provide gentler ionization conditions at the source-MS interface, subambient pressure ionization with nanoelectrospray (SPIN) was utilized. When these improved methods are combined together with the Glycomics Quintavariate Informed Quantification (GlyQ-IQ) recently described (Kronewitter et al. Anal. Chem. 2014, 86, 6268-6276), we are able to significantly extend glycan detection sensitivity and provide expanded glycan coverage. We demonstrated the application of these advances in the context of the human serum glycome, and for which our initial observations included the detection of a new class of heavily sialylated N-glycans, including polysialylated N-glycans.
Figures







Similar articles
-
Improving N-glycan coverage using HPLC-MS with electrospray ionization at subambient pressure.Anal Chem. 2012 Nov 6;84(21):9208-13. doi: 10.1021/ac301961u. Epub 2012 Oct 17. Anal Chem. 2012. PMID: 23025344 Free PMC article.
-
High throughput quantification of N-glycans using one-pot sialic acid modification and matrix assisted laser desorption ionization time-of-flight mass spectrometry.Anal Chem. 2010 Aug 1;82(15):6613-20. doi: 10.1021/ac1011377. Anal Chem. 2010. PMID: 20586471 Free PMC article.
-
Structural analysis of N- and O-glycans released from glycoproteins.Nat Protoc. 2012 Jun 7;7(7):1299-310. doi: 10.1038/nprot.2012.063. Nat Protoc. 2012. PMID: 22678433
-
Structural glycomic analyses at high sensitivity: a decade of progress.Annu Rev Anal Chem (Palo Alto Calif). 2013;6:237-65. doi: 10.1146/annurev-anchem-062012-092609. Epub 2013 Apr 3. Annu Rev Anal Chem (Palo Alto Calif). 2013. PMID: 23560930 Free PMC article. Review.
-
Identification of protein-bound carbohydrates by mass spectrometry.Proteomics. 2001 Feb;1(2):311-28. doi: 10.1002/1615-9861(200102)1:2<311::AID-PROT311>3.0.CO;2-J. Proteomics. 2001. PMID: 11680878 Review.
Cited by
-
A Method for In-Depth Structural Annotation of Human Serum Glycans That Yields Biological Variations.Anal Chem. 2015 Aug 4;87(15):7754-62. doi: 10.1021/acs.analchem.5b01340. Epub 2015 Jul 14. Anal Chem. 2015. PMID: 26086522 Free PMC article.
-
Highly-sensitive label-free deep profiling of N-glycans released from biomedically-relevant samples.Nat Commun. 2023 Mar 23;14(1):1618. doi: 10.1038/s41467-023-37365-4. Nat Commun. 2023. PMID: 36959283 Free PMC article.
-
Nanotechnology in Glycomics: Applications in Diagnostics, Therapy, Imaging, and Separation Processes.Med Res Rev. 2017 May;37(3):514-626. doi: 10.1002/med.21420. Epub 2016 Nov 15. Med Res Rev. 2017. PMID: 27859448 Free PMC article. Review.
-
Recent Advances in the Analysis of Complex Glycoproteins.Anal Chem. 2017 Jan 3;89(1):389-413. doi: 10.1021/acs.analchem.6b04343. Epub 2016 Nov 23. Anal Chem. 2017. PMID: 28105826 Free PMC article. Review. No abstract available.
-
Mass Spectrometry Approaches to Glycomic and Glycoproteomic Analyses.Chem Rev. 2018 Sep 12;118(17):7886-7930. doi: 10.1021/acs.chemrev.7b00732. Epub 2018 Mar 19. Chem Rev. 2018. PMID: 29553244 Free PMC article. Review.
References
-
- Apweiler R.; Hermjakob H.; Sharon N. Biochim. Biophys. Acta: Gen. Subj. 1999, 1473, 4–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

