The protease Omi regulates mitochondrial biogenesis through the GSK3β/PGC-1α pathway
- PMID: 25118933
- PMCID: PMC4454303
- DOI: 10.1038/cddis.2014.328
The protease Omi regulates mitochondrial biogenesis through the GSK3β/PGC-1α pathway
Abstract
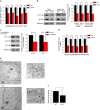
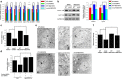
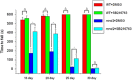
Loss of the mitochondrial protease activity of Omi causes mitochondrial dysfunction, neurodegeneration with parkinsonian features and premature death in mnd2 (motor neuron degeneration 2) mice. However, the detailed mechanisms underlying this pathology remain largely unknown. Here, we report that Omi participates in the process of mitochondrial biogenesis, which has been linked to several neurodegenerative diseases. The mitochondrial biogenesis is deficit in mnd2 mice, evidenced by severe decreases of mitochondrial components, mitochondrial DNA and mitochondrial density. Omi cleaves glycogen synthase kinase 3β (GSK3β), a kinase promoting PPARγ coactivator-1α (PGC-1α) degradation, to regulate PGC-1α, a factor important for the mitochondrial biogenesis. In mnd2 mice, GSK3β abundance is increased and PGC-1α abundance is decreased significantly. Inhibition of GSK3β by SB216763 or overexpression of PGC-1α can restore mitochondrial biogenesis in mnd2 mice or Omi-knockdown N2a cells. Furthermore, there is a significant improvement of the movement ability of mnd2 mice after SB216763 treatment. Thus, our study identified Omi as a novel regulator of mitochondrial biogenesis, involving in Omi protease-deficient-induced neurodegeneration.
Figures




References
-
- Nicholls DG, Budd SL. Mitochondria and neuronal survival. Physiol Rev. 2000;80:315–360. - PubMed
-
- Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. - PubMed
-
- Abou-Sleiman PM, Muqit MM, Wood NW. Expanding insights of mitochondrial dysfunction in Parkinson's disease. Nat Rev Neurosci. 2006;7:207–219. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

