CRISPR-mediated direct mutation of cancer genes in the mouse liver
- PMID: 25119044
- PMCID: PMC4199937
- DOI: 10.1038/nature13589
CRISPR-mediated direct mutation of cancer genes in the mouse liver
Abstract
The study of cancer genes in mouse models has traditionally relied on genetically-engineered strains made via transgenesis or gene targeting in embryonic stem cells. Here we describe a new method of cancer model generation using the CRISPR/Cas (clustered regularly interspaced short palindromic repeats/CRISPR-associated proteins) system in vivo in wild-type mice. We used hydrodynamic injection to deliver a CRISPR plasmid DNA expressing Cas9 and single guide RNAs (sgRNAs) to the liver that directly target the tumour suppressor genes Pten (ref. 5) and p53 (also known as TP53 and Trp53) (ref. 6), alone and in combination. CRISPR-mediated Pten mutation led to elevated Akt phosphorylation and lipid accumulation in hepatocytes, phenocopying the effects of deletion of the gene using Cre-LoxP technology. Simultaneous targeting of Pten and p53 induced liver tumours that mimicked those caused by Cre-loxP-mediated deletion of Pten and p53. DNA sequencing of liver and tumour tissue revealed insertion or deletion mutations of the tumour suppressor genes, including bi-allelic mutations of both Pten and p53 in tumours. Furthermore, co-injection of Cas9 plasmids harbouring sgRNAs targeting the β-catenin gene and a single-stranded DNA oligonucleotide donor carrying activating point mutations led to the generation of hepatocytes with nuclear localization of β-catenin. This study demonstrates the feasibility of direct mutation of tumour suppressor genes and oncogenes in the liver using the CRISPR/Cas system, which presents a new avenue for rapid development of liver cancer models and functional genomics.
Figures


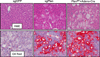


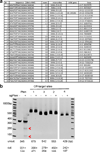


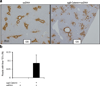



Comment in
-
Cutting short the path to murine liver cancer models.Hepatology. 2015 Jan;61(1):393-5. doi: 10.1002/hep.27580. Epub 2014 Nov 25. Hepatology. 2015. PMID: 25345582 No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
- 2-P01-CA42063/CA/NCI NIH HHS/United States
- R01 CA132091/CA/NCI NIH HHS/United States
- P30 CA014051/CA/NCI NIH HHS/United States
- 5-U54-CA151884-04/CA/NCI NIH HHS/United States
- R01 CA115527/CA/NCI NIH HHS/United States
- R01 DK097768/DK/NIDDK NIH HHS/United States
- R37 EB000244/EB/NIBIB NIH HHS/United States
- 1K99CA169512/CA/NCI NIH HHS/United States
- R01-CA132091/CA/NCI NIH HHS/United States
- R01-EB000244/EB/NIBIB NIH HHS/United States
- R01 EB000244/EB/NIBIB NIH HHS/United States
- R01-CA133404/CA/NCI NIH HHS/United States
- U54 CA151884/CA/NCI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- P01 CA042063/CA/NCI NIH HHS/United States
- P30-CA14051/CA/NCI NIH HHS/United States
- R01-CA115527/CA/NCI NIH HHS/United States
- R01 CA133404/CA/NCI NIH HHS/United States
- DP1 MH100706/MH/NIMH NIH HHS/United States
- K99 CA169512/CA/NCI NIH HHS/United States
- R00 CA169512/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

