Improvements to pairwise sequence comparison (PASC): a genome-based web tool for virus classification
- PMID: 25119676
- PMCID: PMC4221606
- DOI: 10.1007/s00705-014-2197-x
Improvements to pairwise sequence comparison (PASC): a genome-based web tool for virus classification
Abstract
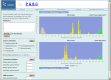
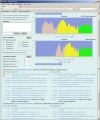
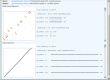
The number of viral genome sequences in the public databases is increasing dramatically, and these sequences are playing an important role in virus classification. Pairwise sequence comparison is a sequence-based virus classification method. A program using this method calculates the pairwise identities of virus sequences within a virus family and displays their distribution, and visual analysis helps to determine demarcations at different taxonomic levels such as strain, species, genus and subfamily. Subsequent comparison of new sequences against existing ones allows viruses from which the new sequences were derived to be classified. Although this method cannot be used as the only criterion for virus classification in some cases, it is a quantitative method and has many advantages over conventional virus classification methods. It has been applied to several virus families, and there is an increasing interest in using this method for other virus families/groups. The Pairwise Sequence Comparison (PASC) classification tool was created at the National Center for Biotechnology Information. The tool's database stores pairwise identities for complete genomes/segments of 56 virus families/groups. Data in the system are updated every day to reflect changes in virus taxonomy and additions of new virus sequences to the public database. The web interface of the tool ( http://www.ncbi.nlm.nih.gov/sutils/pasc/ ) makes it easy to navigate and perform analyses. Multiple new viral genome sequences can be tested simultaneously with this system to suggest the taxonomic position of virus isolates in a specific family. PASC eliminates potential discrepancies in the results caused by different algorithms and/or different data used by researchers.
Figures


References
-
- King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. Virus taxonomy—ninth report of the International Committee on Taxonomy of viruses. London: Elsevier/Academic Press; 2011.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

