The interplay of restriction-modification systems with mobile genetic elements and their prokaryotic hosts
- PMID: 25120263
- PMCID: PMC4176335
- DOI: 10.1093/nar/gku734
The interplay of restriction-modification systems with mobile genetic elements and their prokaryotic hosts
Abstract
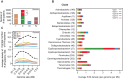
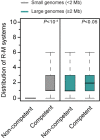
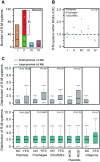
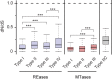
The roles of restriction-modification (R-M) systems in providing immunity against horizontal gene transfer (HGT) and in stabilizing mobile genetic elements (MGEs) have been much debated. However, few studies have precisely addressed the distribution of these systems in light of HGT, its mechanisms and its vectors. We analyzed the distribution of R-M systems in 2261 prokaryote genomes and found their frequency to be strongly dependent on the presence of MGEs, CRISPR-Cas systems, integrons and natural transformation. Yet R-M systems are rare in plasmids, in prophages and nearly absent from other phages. Their abundance depends on genome size for small genomes where it relates with HGT but saturates at two occurrences per genome. Chromosomal R-M systems might evolve under cycles of purifying and relaxed selection, where sequence conservation depends on the biochemical activity and complexity of the system and total gene loss is frequent. Surprisingly, analysis of 43 pan-genomes suggests that solitary R-M genes rarely arise from the degradation of R-M systems. Solitary genes are transferred by large MGEs, whereas complete systems are more frequently transferred autonomously or in small MGEs. Our results suggest means of testing the roles for R-M systems and their associations with MGEs.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Gogarten J.P., Doolittle W.F., Lawrence J.G. Prokaryotic evolution in light of gene transfer. Mol. Biol. Evol. 2002;19:2226–2238. - PubMed
-
- Thomas C.M., Nielsen K.M. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 2005;3:711–721. - PubMed
-
- Labrie S.J., Samson J.E., Moineau S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010;8:317–327. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

