Genome and metabolic network of "Candidatus Phaeomarinobacter ectocarpi" Ec32, a new candidate genus of Alphaproteobacteria frequently associated with brown algae
- PMID: 25120558
- PMCID: PMC4110880
- DOI: 10.3389/fgene.2014.00241
Genome and metabolic network of "Candidatus Phaeomarinobacter ectocarpi" Ec32, a new candidate genus of Alphaproteobacteria frequently associated with brown algae
Abstract
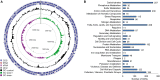
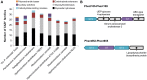
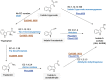
Rhizobiales and related orders of Alphaproteobacteria comprise several genera of nodule-inducing symbiotic bacteria associated with plant roots. Here we describe the genome and the metabolic network of "Candidatus Phaeomarinobacter ectocarpi" Ec32, a member of a new candidate genus closely related to Rhizobiales and found in association with cultures of the filamentous brown algal model Ectocarpus. The "Ca. P. ectocarpi" genome encodes numerous metabolic pathways that may be relevant for this bacterium to interact with algae. Notably, it possesses a large set of glycoside hydrolases and transporters, which may serve to process and assimilate algal metabolites. It also harbors several proteins likely to be involved in the synthesis of algal hormones such as auxins and cytokinins, as well as the vitamins pyridoxine, biotin, and thiamine. As of today, "Ca. P. ectocarpi" has not been successfully cultured, and identical 16S rDNA sequences have been found exclusively associated with Ectocarpus. However, related sequences (≥97% identity) have also been detected free-living and in a Fucus vesiculosus microbiome barcoding project, indicating that the candidate genus "Phaeomarinobacter" may comprise several species, which may colonize different niches.
Keywords: algal-bacterial interactions; genome sequencing; holobiont; metabolic network; phytohormones; symbiosis; transporters; vitamins.
Figures

References
-
- Bartsch I., Wiencke C., Bischof K., Buchholz C. M., Buck B. H., Eggert A., et al. (2008). The genus Laminaria sensu lato: recent insights and developments. Eur. J. Phycol. 43, 1–86 10.1080/09670260701711376 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

