Ebola virus VP24 targets a unique NLS binding site on karyopherin alpha 5 to selectively compete with nuclear import of phosphorylated STAT1
- PMID: 25121748
- PMCID: PMC4188415
- DOI: 10.1016/j.chom.2014.07.008
Ebola virus VP24 targets a unique NLS binding site on karyopherin alpha 5 to selectively compete with nuclear import of phosphorylated STAT1
Abstract
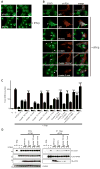
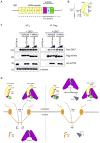
During antiviral defense, interferon (IFN) signaling triggers nuclear transport of tyrosine-phosphorylated STAT1 (PY-STAT1), which occurs via a subset of karyopherin alpha (KPNA) nuclear transporters. Many viruses, including Ebola virus, actively antagonize STAT1 signaling to counteract the antiviral effects of IFN. Ebola virus VP24 protein (eVP24) binds KPNA to inhibit PY-STAT1 nuclear transport and render cells refractory to IFNs. We describe the structure of human KPNA5 C terminus in complex with eVP24. In the complex, eVP24 recognizes a unique nonclassical nuclear localization signal (NLS) binding site on KPNA5 that is necessary for efficient PY-STAT1 nuclear transport. eVP24 binds KPNA5 with very high affinity to effectively compete with and inhibit PY-STAT1 nuclear transport. In contrast, eVP24 binding does not affect the transport of classical NLS cargo. Thus, eVP24 counters cell-intrinsic innate immunity by selectively targeting PY-STAT1 nuclear import while leaving the transport of other cargo that may be required for viral replication unaffected.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures




Comment in
-
How a virus blocks a cellular emergency access lane to the nucleus, STAT!Cell Host Microbe. 2014 Aug 13;16(2):150-152. doi: 10.1016/j.chom.2014.07.013. Cell Host Microbe. 2014. PMID: 25121743
References
-
- Bray M, Murphy FA. Filovirus research: knowledge expands to meet a growing threat. J Infect Dis. 2007;196(Suppl 2):S438–443. - PubMed
-
- Chen X, Vinkemeier U, Zhao Y, Jeruzalmi D, Darnell JE, Jr, Kuriyan J. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell. 1998;93:827–839. - PubMed
-
- Chook YM, Blobel G. Karyopherins and nuclear import. Curr Opin Struct Biol. 2001;11:703–715. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- R01HL119813/HL/NHLBI NIH HHS/United States
- R01 AI081914/AI/NIAID NIH HHS/United States
- R01 AI107056/AI/NIAID NIH HHS/United States
- U19 AI070489/AI/NIAID NIH HHS/United States
- U19AI109664/AI/NIAID NIH HHS/United States
- T32GM07200/GM/NIGMS NIH HHS/United States
- R01AI059536/AI/NIAID NIH HHS/United States
- R01 AI059536/AI/NIAID NIH HHS/United States
- U19 AI109945/AI/NIAID NIH HHS/United States
- U19AI109945/AI/NIAID NIH HHS/United States
- R01AI107056/AI/NIAID NIH HHS/United States
- P41 GM103399/GM/NIGMS NIH HHS/United States
- R01AI081914/AI/NIAID NIH HHS/United States
- R01 HL119813/HL/NHLBI NIH HHS/United States
- T32 HL007317/HL/NHLBI NIH HHS/United States
- U19AI070489/AI/NIAID NIH HHS/United States
- U19 AI109664/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

