MADS-box transcription factor AGL21 regulates lateral root development and responds to multiple external and physiological signals
- PMID: 25122697
- PMCID: PMC4228986
- DOI: 10.1093/mp/ssu088
MADS-box transcription factor AGL21 regulates lateral root development and responds to multiple external and physiological signals
Abstract
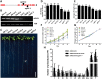
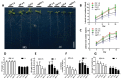
Plant root system morphology is dramatically influenced by various environmental cues. The adaptation of root system architecture to environmental constraints, which mostly depends on the formation and growth of lateral roots, is an important agronomic trait. Lateral root development is regulated by the external signals coordinating closely with intrinsic signaling pathways. MADS-box transcription factors are known key regulators of the transition to flowering and flower development. However, their functions in root development are still poorly understood. Here we report that AGL21, an AGL17-clade MADS-box gene, plays a crucial role in lateral root development. AGL21 was highly expressed in root, particularly in the root central cylinder and lateral root primordia. AGL21 overexpression plants produced more and longer lateral roots while agl21 mutants showed impaired lateral root development, especially under nitrogen-deficient conditions. AGL21 was induced by many plant hormones and environmental stresses, suggesting a function of this gene in root system plasticity in response to various signals. Furthermore, AGL21 was found positively regulating auxin accumulation in lateral root primordia and lateral roots by enhancing local auxin biosynthesis, thus stimulating lateral root initiation and growth. We propose that AGL21 may be involved in various environmental and physiological signals-mediated lateral root development and growth.
Keywords: AGL21; MADS; auxin; lateral root; nitrate; root system architecture; sulfate..
© The Author 2014. Published by Oxford University Press on behalf of CSPB and IPPE, SIBS, CAS.
Figures








References
-
- Achard P., Cheng H., De Grauwe L., Decat J., Schoutteten H., Moritz T., Van Der Straeten D., Peng J., Harberd N.P. (2006). Integration of plant responses to environmentally activated phytohormonal signals. Science. 311, 91–94. - PubMed
-
- Alvarez-Buylla E.R., Liljegren S.J., Pelaz S., Gold S.E., Burgeff C., Ditta G.S., Vergara-Silva F., Yanofsky M.F. (2000a). MADS-box gene evolution beyond flowers: expression in pollen, endosperm, guard cells, roots and trichomes. Plant J. 24, 457–466. - PubMed
-
- Alvarez-Buylla E.R., Pelaz S., Liljegren S.J., Gold S.E., Burgeff C., Ditta G.S., Ribas de Pouplana L., Martinez-Castilla L., Yanofsky M.F. (2000b). An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl Acad. Sci. U S A. 97, 5328–5333. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

