Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector
- PMID: 25122746
- PMCID: PMC4231726
- DOI: 10.1093/nar/gku749
Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector
Abstract
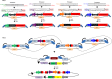
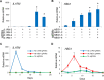
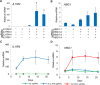
Engineered DNA-binding proteins that manipulate the human genome and transcriptome have enabled rapid advances in biomedical research. In particular, the RNA-guided CRISPR/Cas9 system has recently been engineered to create site-specific double-strand breaks for genome editing or to direct targeted transcriptional regulation. A unique capability of the CRISPR/Cas9 system is multiplex genome engineering by delivering a single Cas9 enzyme and two or more single guide RNAs (sgRNAs) targeted to distinct genomic sites. This approach can be used to simultaneously create multiple DNA breaks or to target multiple transcriptional activators to a single promoter for synergistic enhancement of gene induction. To address the need for uniform and sustained delivery of multiplex CRISPR/Cas9-based genome engineering tools, we developed a single lentiviral system to express a Cas9 variant, a reporter gene and up to four sgRNAs from independent RNA polymerase III promoters that are incorporated into the vector by a convenient Golden Gate cloning method. Each sgRNA is efficiently expressed and can mediate multiplex gene editing and sustained transcriptional activation in immortalized and primary human cells. This delivery system will be significant to enabling the potential of CRISPR/Cas9-based multiplex genome engineering in diverse cell types.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Urnov F.D., Miller J.C., Lee Y.L., Beausejour C.M., Rock J.M., Augustus S., Jamieson A.C., Porteus M.H., Gregory P.D., Holmes M.C. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

