Molecular insights into replication initiation by Qβ replicase using ribosomal protein S1
- PMID: 25122749
- PMCID: PMC4176380
- DOI: 10.1093/nar/gku745
Molecular insights into replication initiation by Qβ replicase using ribosomal protein S1
Abstract
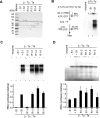
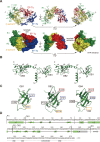
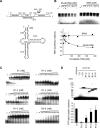
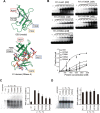
Ribosomal protein S1, consisting of six contiguous OB-folds, is the largest ribosomal protein and is essential for translation initiation in Escherichia coli. S1 is also one of the three essential host-derived subunits of Qβ replicase, together with EF-Tu and EF-Ts, for Qβ RNA replication in E. coli. We analyzed the crystal structure of Qβ replicase, consisting of the virus-encoded RNA-dependent RNA polymerase (β-subunit), EF-Tu, EF-Ts and the N-terminal half of S1, which is capable of initiating Qβ RNA replication. Structural and biochemical studies revealed that the two N-terminal OB-folds of S1 anchor S1 onto the β-subunit, and the third OB-fold is mobile and protrudes beyond the surface of the β-subunit. The third OB-fold mainly interacts with a specific RNA fragment derived from the internal region of Qβ RNA, and its RNA-binding ability is required for replication initiation of Qβ RNA. Thus, the third mobile OB-fold of S1, which is spatially anchored near the surface of the β-subunit, primarily recruits the Qβ RNA toward the β-subunit, leading to the specific and efficient replication initiation of Qβ RNA, and S1 functions as a replication initiation factor, beyond its established function in protein synthesis.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Subramanian A.R. Structure and functions of ribosomal protein S1. Prog. Nucleic Acid Res. Mol. Biol. 1983;28:102–138. - PubMed
-
- Farwell M.A., Robert M.W., Rabinowitz J.C. The effect of ribosomal protein S1 from Escherichia coli and Micrococcus luteus on protein synthesis in vitro by E. coli and Bacillus subtilis. Mol. Microbiol. 1992;6:3375–3383. - PubMed
-
- Tedin K., Resch A., Blasi U. Requirements for ribosomal protein S1 for translation initiation of mRNAs with and without a 5′ leader sequence. Mol. Microbiol. 1997;25:189–199. - PubMed
-
- Sorensen M.A., Fricke J., Pedersen S. Ribosomal protein S1 is required for translation of most, if not all, natural mRNAs in Escherichia coli in vivo. J. Mol. Biol. 1998;280:561–569. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

