Characterization of transcriptomes of cochlear inner and outer hair cells
- PMID: 25122905
- PMCID: PMC4131018
- DOI: 10.1523/JNEUROSCI.1690-14.2014
Characterization of transcriptomes of cochlear inner and outer hair cells
Abstract
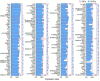
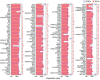
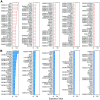
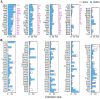
Inner hair cells (IHCs) and outer hair cells (OHCs) are the two types of sensory receptor cells that are critical for hearing in the mammalian cochlea. IHCs and OHCs have different morphology and function. The genetic mechanisms that define their morphological and functional specializations are essentially unknown. The transcriptome reflects the genes that are being actively expressed in a cell and holds the key to understanding the molecular mechanisms of the biological properties of the cell. Using DNA microarray, we examined the transcriptome of 2000 individually collected IHCs and OHCs from adult mouse cochleae. We show that 16,647 and 17,711 transcripts are expressed in IHCs and OHCs, respectively. Of those genes, ∼73% are known genes, 22% are uncharacterized sequences, and 5.0% are noncoding RNAs in both populations. A total of 16,117 transcripts are expressed in both populations. Uniquely and differentially expressed genes account for <15% of all genes in either cell type. The top 10 differentially expressed genes include Slc17a8, Dnajc5b, Slc1a3, Atp2a3, Osbpl6, Slc7a14, Bcl2, Bin1, Prkd1, and Map4k4 in IHCs and Slc26a5, C1ql1, Strc, Dnm3, Plbd1, Lbh, Olfm1, Plce1, Tectb, and Ankrd22 in OHCs. We analyzed commonly and differentially expressed genes with the focus on genes related to hair cell specializations in the apical, basolateral, and synaptic membranes. Eighty-three percent of the known deafness-related genes are expressed in hair cells. We also analyzed genes involved in cell-cycle regulation. Our dataset holds an extraordinary trove of information about the molecular mechanisms underlying hair cell morphology, function, pathology, and cell-cycle control.
Keywords: DNA microarray; inner hair cells; mouse; outer hair cells; transcriptome.
Copyright © 2014 the authors 0270-6474/14/3411085-11$15.00/0.
Figures



Similar articles
-
Transcriptomes of cochlear inner and outer hair cells from adult mice.Sci Data. 2018 Oct 2;5:180199. doi: 10.1038/sdata.2018.199. Sci Data. 2018. PMID: 30277483 Free PMC article.
-
Trans-differentiation of outer hair cells into inner hair cells in the absence of INSM1.Nature. 2018 Nov;563(7733):691-695. doi: 10.1038/s41586-018-0570-8. Epub 2018 Oct 10. Nature. 2018. PMID: 30305733 Free PMC article.
-
Transcription Factors Expressed in Mouse Cochlear Inner and Outer Hair Cells.PLoS One. 2016 Mar 14;11(3):e0151291. doi: 10.1371/journal.pone.0151291. eCollection 2016. PLoS One. 2016. PMID: 26974322 Free PMC article.
-
Recent advances in molecular studies on cochlear development and regeneration.Curr Opin Neurobiol. 2023 Aug;81:102745. doi: 10.1016/j.conb.2023.102745. Epub 2023 Jun 23. Curr Opin Neurobiol. 2023. PMID: 37356371 Review.
-
Two types of cochlear hair cells with two different modes of activation are better than one.J Basic Clin Physiol Pharmacol. 2012 Jan 11;23(1):1-3. doi: 10.1515/jbcpp-2011-0036. J Basic Clin Physiol Pharmacol. 2012. PMID: 22865443 Review.
Cited by
-
Quantitative Fluorescent in situ Hybridization Reveals Differential Transcription Profile Sharpening of Endocytic Proteins in Cochlear Hair Cells Upon Maturation.Front Cell Neurosci. 2021 Feb 26;15:643517. doi: 10.3389/fncel.2021.643517. eCollection 2021. Front Cell Neurosci. 2021. PMID: 33716676 Free PMC article.
-
Assessment of the expression and role of the α1-nAChR subunit in efferent cholinergic function during the development of the mammalian cochlea.J Neurophysiol. 2016 Aug 1;116(2):479-92. doi: 10.1152/jn.01038.2015. Epub 2016 Apr 20. J Neurophysiol. 2016. PMID: 27098031 Free PMC article.
-
Dual role for Sox2 in specification of sensory competence and regulation of Atoh1 function.Dev Neurobiol. 2017 Jan;77(1):3-13. doi: 10.1002/dneu.22401. Epub 2016 Jun 6. Dev Neurobiol. 2017. PMID: 27203669 Free PMC article.
-
Auditory Nomenclature: Combining Name Recognition With Anatomical Description.Front Neuroanat. 2018 Nov 23;12:99. doi: 10.3389/fnana.2018.00099. eCollection 2018. Front Neuroanat. 2018. PMID: 30532697 Free PMC article. Review.
-
Identification of two novel mutations in POU4F3 gene associated with autosomal dominant hearing loss in Chinese families.J Cell Mol Med. 2020 Jun;24(12):6978-6987. doi: 10.1111/jcmm.15359. Epub 2020 May 11. J Cell Mol Med. 2020. PMID: 32390314 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
