Amyloid precursor protein dimerization and synaptogenic function depend on copper binding to the growth factor-like domain
- PMID: 25122912
- PMCID: PMC6705248
- DOI: 10.1523/JNEUROSCI.0180-14.2014
Amyloid precursor protein dimerization and synaptogenic function depend on copper binding to the growth factor-like domain
Abstract
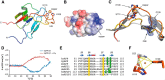
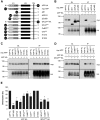
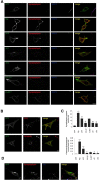
Accumulating evidence suggests that the copper-binding amyloid precursor protein (APP) has an essential synaptic function. APP synaptogenic function depends on trans-directed dimerization of the extracellular E1 domain encompassing a growth factor-like domain (GFLD) and a copper-binding domain (CuBD). Here we report the 1.75 Å crystal structure of the GFLD in complex with a copper ion bound with high affinity to an extended hairpin loop at the dimerization interface. In coimmunoprecipitation assays copper binding promotes APP interaction, whereas mutations in the copper-binding sites of either the GFLD or CuBD result in a drastic reduction in APP cis-orientated dimerization. We show that copper is essential and sufficient to induce trans-directed dimerization of purified APP. Furthermore, a mixed culture assay of primary neurons with HEK293 cells expressing different APP mutants revealed that APP potently promotes synaptogenesis depending on copper binding to the GFLD. Together, these findings demonstrate that copper binding to the GFLD of APP is required for APP cis-/trans-directed dimerization and APP synaptogenic function. Thus, neuronal activity or disease-associated changes in copper homeostasis likely go along with altered APP synaptic function.
Keywords: Alzheimer's disease; amyloid precursor protein; copper; dimerization; metal homeostasis; synaptogenesis.
Copyright © 2014 the authors 0270-6474/14/3411159-14$15.00/0.
Figures





Similar articles
-
Copper and zinc ions govern the trans-directed dimerization of APP family members in multiple ways.J Neurochem. 2019 Dec;151(5):626-641. doi: 10.1111/jnc.14716. Epub 2019 Jun 6. J Neurochem. 2019. PMID: 31063592
-
Copper enhances APP dimerization and promotes Aβ production.Neurosci Lett. 2013 Jun 28;547:10-5. doi: 10.1016/j.neulet.2013.04.057. Epub 2013 May 11. Neurosci Lett. 2013. PMID: 23669644
-
Structure of the Alzheimer's disease amyloid precursor protein copper binding domain. A regulator of neuronal copper homeostasis.J Biol Chem. 2003 May 9;278(19):17401-7. doi: 10.1074/jbc.M300629200. Epub 2003 Feb 28. J Biol Chem. 2003. PMID: 12611883
-
Copper binding to the Alzheimer's disease amyloid precursor protein.Eur Biophys J. 2008 Mar;37(3):269-79. doi: 10.1007/s00249-007-0234-3. Epub 2007 Nov 21. Eur Biophys J. 2008. PMID: 18030462 Free PMC article. Review.
-
Copper brain homeostasis: role of amyloid precursor protein and prion protein.IUBMB Life. 2005 Sep;57(9):645-50. doi: 10.1080/15216540500264620. IUBMB Life. 2005. PMID: 16203684 Review.
Cited by
-
The Brilliance of the Zebrafish Model: Perception on Behavior and Alzheimer's Disease.Front Behav Neurosci. 2022 Jun 13;16:861155. doi: 10.3389/fnbeh.2022.861155. eCollection 2022. Front Behav Neurosci. 2022. PMID: 35769627 Free PMC article. Review.
-
Neurotoxicity Linked to Dysfunctional Metal Ion Homeostasis and Xenobiotic Metal Exposure: Redox Signaling and Oxidative Stress.Antioxid Redox Signal. 2018 Jun 20;28(18):1669-1703. doi: 10.1089/ars.2017.7272. Epub 2018 Mar 28. Antioxid Redox Signal. 2018. PMID: 29402131 Free PMC article. Review.
-
Amyloid precursor protein and its interacting proteins in neurodevelopment.Biochem Soc Trans. 2023 Aug 31;51(4):1647-1659. doi: 10.1042/BST20221527. Biochem Soc Trans. 2023. PMID: 37387352 Free PMC article. Review.
-
Shedding of APP limits its synaptogenic activity and cell adhesion properties.Front Cell Neurosci. 2014 Dec 3;8:410. doi: 10.3389/fncel.2014.00410. eCollection 2014. Front Cell Neurosci. 2014. PMID: 25520622 Free PMC article.
-
The Impact of Biomaterial Cell Contact on the Immunopeptidome.Front Bioeng Biotechnol. 2020 Dec 16;8:571294. doi: 10.3389/fbioe.2020.571294. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 33392160 Free PMC article.
References
-
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
