Genetic diversity of porcine group A rotavirus strains in the UK
- PMID: 25123085
- PMCID: PMC4158422
- DOI: 10.1016/j.vetmic.2014.06.030
Genetic diversity of porcine group A rotavirus strains in the UK
Erratum in
- Vet Microbiol. 2014 Dec 5;174(3-4):609. Mellits, Kenneth M [corrected to Mellits, Kenneth H]
Abstract
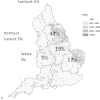
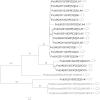
Rotavirus is endemic in pig farms where it causes a loss in production. This study is the first to characterise porcine rotavirus circulating in UK pigs. Samples from diarrheic pigs with rotavirus enteritis obtained between 2010 and 2012 were genotyped in order to determine the diversity of group A rotavirus (GARV) in UK pigs. A wide range of rotavirus genotypes were identified in UK pigs: six G types (VP7); G2, G3, G4, G5, G9 and G11 and six P types (VP4); P[6], P[7], P[8], P[13], P[23], and P[32]. With the exception of a single P[8] isolate, there was less than 95% nucleotide identity between sequences from this study and any available rotavirus sequences. The G9 and P[6] genotypes are capable of infecting both humans and pigs, but showed no species cross-over within the UK as they were shown to be genetically distinct, which suggested zoonotic transmission is rare within the UK. We identified the P[8] genotype in one isolate, this genotype is almost exclusively found in humans. The P[8] was linked to a human Irish rotavirus isolate in the same year. The discovery of human genotype P[8] rotavirus in a UK pig confirms this common human genotype can infect pigs and also highlights the necessity of surveillance of porcine rotavirus genotypes to safeguard human as well as porcine health.
Keywords: Phylogenetic; Porcine; Rotavirus; Zoonosis.
Copyright © 2014. Published by Elsevier B.V.
Figures




References
-
- Bhan M.K., Lew J.F., Sazawal S., Das B.K., Gentsch J.R., Glass R.I. Protection conferred by neonatal rotavirus infection against subsequent rotavirus diarrhea. J. Infect. Dis. 1993;168:282–287. - PubMed
-
- Cashman O., Collins P.J., Lennon G., Cryan B., Martella V., Fanning S., Staines A., O'Shea H. Molecular characterization of group A rotaviruses detected in children with gastroenteritis in Ireland in 2006–2009. Epidemiol. Infect. 2012;140:247–259. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

