Deep sequencing reveals clonal evolution patterns and mutation events associated with relapse in B-cell lymphomas
- PMID: 25123191
- PMCID: PMC4158101
- DOI: 10.1186/s13059-014-0432-0
Deep sequencing reveals clonal evolution patterns and mutation events associated with relapse in B-cell lymphomas
Abstract
Background: Molecular mechanisms associated with frequent relapse of diffuse large B-cell lymphoma (DLBCL) are poorly defined. It is especially unclear how primary tumor clonal heterogeneity contributes to relapse. Here, we explore unique features of B-cell lymphomas - VDJ recombination and somatic hypermutation - to address this question.
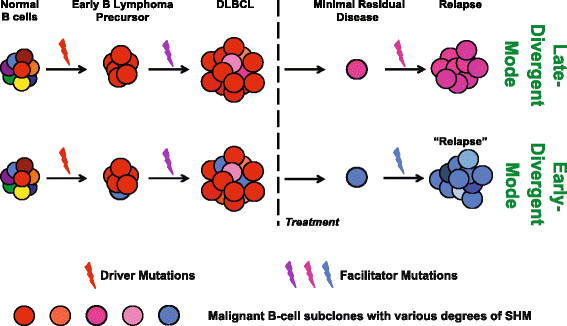
Results: We performed high-throughput sequencing of rearranged VDJ junctions in 14 pairs of matched diagnosis-relapse tumors, among which 7 pairs were further characterized by exome sequencing. We identify two distinctive modes of clonal evolution of DLBCL relapse: an early-divergent mode in which clonally related diagnosis and relapse tumors diverged early and developed in parallel; and a late-divergent mode in which relapse tumors developed directly from diagnosis tumors with minor divergence. By examining mutation patterns in the context of phylogenetic information provided by VDJ junctions, we identified mutations in epigenetic modifiers such as KMT2D as potential early driving events in lymphomagenesis and immune escape alterations as relapse-associated events.
Conclusions: Altogether, our study for the first time provides important evidence that DLBCL relapse may result from multiple, distinct tumor evolutionary mechanisms, providing rationale for therapies for each mechanism. Moreover, this study highlights the urgent need to understand the driving roles of epigenetic modifier mutations in lymphomagenesis, and immune surveillance factor genetic lesions in relapse.
Figures





References
-
- Coiffier B, Lepage E, Briere J, Herbrecht R, Tilly H, Bouabdallah R, Morel P, Van Den Neste E, Salles G, Gaulard P, Reyes F, Lederlin P, Gisselbrecht C. CHOP chemotherapy plus rituximab compared with CHOP alone in elderly patients with diffuse large-B-cell lymphoma. N Engl J Med. 2002;346:235–242. doi: 10.1056/NEJMoa011795. - DOI - PubMed
-
- Larouche JF, Berger F, Chassagne-Clement C, Ffrench M, Callet-Bauchu E, Sebban C, Ghesquieres H, Broussais-Guillaumot F, Salles G, Coiffier B. Lymphoma recurrence 5 years or later following diffuse large B-cell lymphoma: clinical characteristics and outcome. J Clin Oncol. 2010;28:2094–2100. doi: 10.1200/JCO.2009.24.5860. - DOI - PubMed
-
- Gisselbrecht C, Glass B, Mounier N, Singh Gill D, Linch DC, Trneny M, Bosly A, Ketterer N, Shpilberg O, Hagberg H, Ma D, Briere J, Moskowitz CH, Schmitz N. Salvage regimens with autologous transplantation for relapsed large B-cell lymphoma in the rituximab era. J Clin Oncol. 2010;28:4184–4190. doi: 10.1200/JCO.2010.28.1618. - DOI - PMC - PubMed
-
- Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, Ritchey JK, Young MA, Lamprecht T, McLellan MD, McMichael JF, Wallis JW, Lu C, Shen D, Harris CC, Dooling DJ, Fulton RS, Fulton LL, Chen K, Schmidt H, Kalicki-Veizer J, Magrini VJ, Cook L, McGrath SD, Vickery TL, Wendl MC, Heath S, Watson MA, Link DC, Tomasson MH, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–510. doi: 10.1038/nature10738. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

