Considerations when investigating lncRNA function in vivo
- PMID: 25124674
- PMCID: PMC4132285
- DOI: 10.7554/eLife.03058
Considerations when investigating lncRNA function in vivo
Abstract
Although a small number of the vast array of animal long non-coding RNAs (lncRNAs) have known effects on cellular processes examined in vitro, the extent of their contributions to normal cell processes throughout development, differentiation and disease for the most part remains less clear. Phenotypes arising from deletion of an entire genomic locus cannot be unequivocally attributed either to the loss of the lncRNA per se or to the associated loss of other overlapping DNA regulatory elements. The distinction between cis- or trans-effects is also often problematic. We discuss the advantages and challenges associated with the current techniques for studying the in vivo function of lncRNAs in the light of different models of lncRNA molecular mechanism, and reflect on the design of experiments to mutate lncRNA loci. These considerations should assist in the further investigation of these transcriptional products of the genome.
Keywords: Science forum; brain development; developmental defect; knockout mouse models; lethality; long non-coding RNAs.
Copyright © 2014, Bassett et al.
Conflict of interest statement
CPP: Senior Editor,
AA: Reviewing Editor,
ACF-S: Reviewing Editor,
TRG: Reviewing Editor,
The other authors declare that no competing interests exist.
Figures
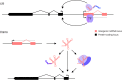


References
Publication types
MeSH terms
Substances
Grants and funding
- MC_UU_12021/1/MRC_/Medical Research Council/United Kingdom
- U54 HG007004/HG/NHGRI NIH HHS/United States
- 095606/WT_/Wellcome Trust/United Kingdom
- MC_U137761446/MRC_/Medical Research Council/United Kingdom
- U54 HG007004-2./HG/NHGRI NIH HHS/United States
- G1000801B/MRC_/Medical Research Council/United Kingdom
- P30 CA045508/CA/NCI NIH HHS/United States
- G1000801/MRC_/Medical Research Council/United Kingdom
- MC_UU_12009/4/MRC_/Medical Research Council/United Kingdom
- 092076/WT_/Wellcome Trust/United Kingdom
- 11832/CRUK_/Cancer Research UK/United Kingdom
- 249869/ERC_/European Research Council/International
- 103768/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

