The regulation of coenzyme q biosynthesis in eukaryotic cells: all that yeast can tell us
- PMID: 25126044
- PMCID: PMC4112530
- DOI: 10.1159/000362897
The regulation of coenzyme q biosynthesis in eukaryotic cells: all that yeast can tell us
Abstract
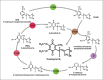
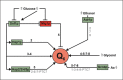
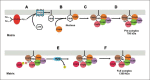
Coenzyme Q (CoQ) is a mitochondrial lipid, which functions mainly as an electron carrier from complex I or II to complex III at the mitochondrial inner membrane, and also as antioxidant in cell membranes. CoQ is needed as electron acceptor in β-oxidation of fatty acids and pyridine nucleotide biosynthesis, and it is responsible for opening the mitochondrial permeability transition pore. The yeast model has been very useful to analyze the synthesis of CoQ, and therefore, most of the knowledge about its regulation was obtained from the Saccharomyces cerevisiae model. CoQ biosynthesis is regulated to support 2 processes: the bioenergetic metabolism and the antioxidant defense. Alterations of the carbon source in yeast, or in nutrient availability in yeasts or mammalian cells, upregulate genes encoding proteins involved in CoQ synthesis. Oxidative stress, generated by chemical or physical agents or by serum deprivation, modifies specifically the expression of some COQ genes by means of stress transcription factors such as Msn2/4p, Yap1p or Hsf1p. In general, the induction of COQ gene expression produced by metabolic changes or stress is modulated downstream by other regulatory mechanisms such as the protein import to mitochondria, the assembly of a multi-enzymatic complex composed by Coq proteins and also the existence of a phosphorylation cycle that regulates the last steps of CoQ biosynthesis. The CoQ biosynthetic complex assembly starts with the production of a nucleating lipid such as HHB by the action of the Coq2 protein. Then, the Coq4 protein recognizes the precursor HHB acting as the nucleus of the complex. The activity of Coq8p, probably as kinase, allows the formation of an initial pre-complex containing all Coq proteins with the exception of Coq7p. This pre-complex leads to the synthesis of 5-demethoxy-Q6 (DMQ6), the Coq7p substrate. When de novo CoQ biosynthesis is required, Coq7p becomes dephosphorylated by the action of Ptc7p increasing the synthesis rate of CoQ6. This critical model is needed for a better understanding of CoQ biosynthesis. Taking into account that patients with CoQ10 deficiency maintain to some extent the machinery to synthesize CoQ, new promising strategies for the treatment of CoQ10 deficiency will require a better understanding of the regulation of CoQ biosynthesis in the future.
Keywords: Coenzyme Q; Mitochondria; Protein complex; Respiration; Ubiquinone; Yeast.
Figures
Similar articles
-
Regulation of coenzyme Q biosynthesis in yeast: a new complex in the block.IUBMB Life. 2014 Feb;66(2):63-70. doi: 10.1002/iub.1243. Epub 2014 Jan 27. IUBMB Life. 2014. PMID: 24470391 Review.
-
Coenzyme Q supplementation or over-expression of the yeast Coq8 putative kinase stabilizes multi-subunit Coq polypeptide complexes in yeast coq null mutants.Biochim Biophys Acta. 2014 Apr 4;1841(4):630-44. doi: 10.1016/j.bbalip.2013.12.017. Epub 2014 Jan 7. Biochim Biophys Acta. 2014. PMID: 24406904 Free PMC article.
-
Respiratory-induced coenzyme Q biosynthesis is regulated by a phosphorylation cycle of Cat5p/Coq7p.Biochem J. 2011 Nov 15;440(1):107-14. doi: 10.1042/BJ20101422. Biochem J. 2011. PMID: 21812761
-
COQ11 deletion mitigates respiratory deficiency caused by mutations in the gene encoding the coenzyme Q chaperone protein Coq10.J Biol Chem. 2020 May 1;295(18):6023-6042. doi: 10.1074/jbc.RA119.012420. Epub 2020 Mar 23. J Biol Chem. 2020. PMID: 32205446 Free PMC article.
-
Coenzyme Q10 deficiencies: pathways in yeast and humans.Essays Biochem. 2018 Jul 20;62(3):361-376. doi: 10.1042/EBC20170106. Print 2018 Jul 20. Essays Biochem. 2018. PMID: 29980630 Free PMC article. Review.
Cited by
-
Optogenetic control of horizontally acquired genes prevent stuck fermentations in yeast.Microbiol Spectr. 2025 Feb 4;13(2):e0179424. doi: 10.1128/spectrum.01794-24. Epub 2025 Jan 8. Microbiol Spectr. 2025. PMID: 39772912 Free PMC article.
-
RNA-binding proteins regulate cell respiration and coenzyme Q biosynthesis by post-transcriptional regulation of COQ7.RNA Biol. 2016 Jul 2;13(7):622-34. doi: 10.1080/15476286.2015.1119366. Epub 2015 Dec 21. RNA Biol. 2016. PMID: 26690054 Free PMC article.
-
Biochemistry of Mitochondrial Coenzyme Q Biosynthesis.Trends Biochem Sci. 2017 Oct;42(10):824-843. doi: 10.1016/j.tibs.2017.06.008. Epub 2017 Sep 17. Trends Biochem Sci. 2017. PMID: 28927698 Free PMC article. Review.
-
Chromatin-remodeling SWI/SNF complex regulates coenzyme Q6 synthesis and a metabolic shift to respiration in yeast.J Biol Chem. 2017 Sep 8;292(36):14851-14866. doi: 10.1074/jbc.M117.798397. Epub 2017 Jul 24. J Biol Chem. 2017. PMID: 28739803 Free PMC article.
-
The UPRmt Protects Caenorhabditis elegans from Mitochondrial Dysfunction by Upregulating Specific Enzymes of the Mevalonate Pathway.Genetics. 2018 Jun;209(2):457-473. doi: 10.1534/genetics.118.300863. Epub 2018 Mar 29. Genetics. 2018. PMID: 29599115 Free PMC article.
References
-
- Antonenkov VD, Grunau S, Ohlmeier S, Hiltunen JK. Peroxisomes are oxidative organelles. Antioxid Redox Signal. 2010;13:525–537. - PubMed
-
- Arroyo BA, Villalba JMJM, Arroyo A, Rodríguez-Aguilera JCJ, Santos-Ocaña C, Navas P. Stabilization of extracellular ascorbate mediated by coenzyme Q transmembrane electron transport. Methods Enzymol. 2004;378:207–217. - PubMed
-
- Artuch R, Brea-Calvo G, Briones P, Aracil A, Galvan M, et al. Cerebellar ataxia with coenzyme Q10 deficiency: diagnosis and follow-up after coenzyme Q10 supplementation. J Neurol Sci. 2006;246:153–158. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

