Metabolic phenotyping reveals a lipid mediator response to ionizing radiation
- PMID: 25126707
- PMCID: PMC4156265
- DOI: 10.1021/pr5005295
Metabolic phenotyping reveals a lipid mediator response to ionizing radiation
Abstract
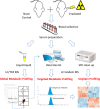
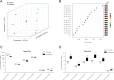
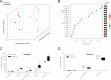
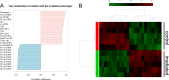
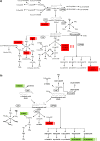
Exposure to ionizing radiation has dramatically increased in modern society, raising serious health concerns. The molecular response to ionizing radiation, however, is still not completely understood. Here, we screened mouse serum for metabolic alterations following an acute exposure to γ radiation using a multiplatform mass-spectrometry-based strategy. A global, molecular profiling revealed that mouse serum undergoes a series of significant molecular alterations following radiation exposure. We identified and quantified bioactive metabolites belonging to key biochemical pathways and low-abundance, oxygenated, polyunsaturated fatty acids (PUFAs) in the two groups of animals. Exposure to γ radiation induced a significant increase in the serum levels of ether phosphatidylcholines (PCs) while decreasing the levels of diacyl PCs carrying PUFAs. In exposed mice, levels of pro-inflammatory, oxygenated metabolites of arachidonic acid increased, whereas levels of anti-inflammatory metabolites of omega-3 PUFAs decreased. Our results indicate a specific serum lipidomic biosignature that could be utilized as an indicator of radiation exposure and as novel target for therapeutic intervention. Monitoring such a molecular response to radiation exposure might have implications not only for radiation pathology but also for countermeasures and personalized medicine.
Figures
References
-
- Brenner D. J.; Hall E. J. Computed tomography—an increasing source of radiation exposure. N. Engl. J. Med. 2007, 357, 2277–84. - PubMed
-
- Pearce M. S.; Salotti J. A.; Little M. P.; McHugh K.; Lee C.; Kim K. P.; Howe N. L.; Ronckers C. M.; Rajaraman P.; Sir Craft A. W.; Parker L.; Berrington de Gonzalez A. Radiation exposure from CT scans in childhood and subsequent risk of leukaemia and brain tumours: a retrospective cohort study. Lancet 2012, 380, 499–505. - PMC - PubMed
-
- Mathews J. D.; Forsythe A. V.; Brady Z.; Butler M. W.; Goergen S. K.; Byrnes G. B.; Giles G. G.; Wallace A. B.; Anderson P. R.; Guiver T. A.; McGale P.; Cain T. M.; Dowty J. G.; Bickerstaffe A. C.; Darby S. C. Cancer risk in 680,000 people exposed to computed tomography scans in childhood or adolescence: data linkage study of 11 million Australians. BMJ 2013, 346, f2360. - PMC - PubMed
-
- Miglioretti D. L.; Johnson E.; Williams A.; Greenlee R. T.; Weinmann S.; Solberg L. I.; Feigelson H. S.; Roblin D.; Flynn M. J.; Vanneman N.; Smith-Bindman R. The use of computed tomography in pediatrics and the associated radiation exposure and estimated cancer risk. JAMA Pediatr 2013, 167, 700–7. - PMC - PubMed
-
- Ozasa K.; Shimizu Y.; Sakata R.; Sugiyama H.; Grant E. J.; Soda M.; Kasagi F.; Suyama A. Risk of cancer and non-cancer diseases in the atomic bomb survivors. Radiat. Prot. Dosim. 2011, 146, 272–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

