Function of a Foxp3 cis-element in protecting regulatory T cell identity
- PMID: 25126782
- PMCID: PMC4151505
- DOI: 10.1016/j.cell.2014.07.030
Function of a Foxp3 cis-element in protecting regulatory T cell identity
Abstract
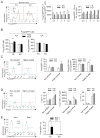
The homeostasis of multicellular organisms requires terminally differentiated cells to preserve their lineage specificity. However, it is unclear whether mechanisms exist to actively protect cell identity in response to environmental cues that confer functional plasticity. Regulatory T (Treg) cells, specified by the transcription factor Foxp3, are indispensable for immune system homeostasis. Here, we report that conserved noncoding sequence 2 (CNS2), a CpG-rich Foxp3 intronic cis-element specifically demethylated in mature Tregs, helps maintain immune homeostasis and limit autoimmune disease development by protecting Treg identity in response to signals that shape mature Treg functions and drive their initial differentiation. In activated Tregs, CNS2 helps protect Foxp3 expression from destabilizing cytokine conditions by sensing TCR/NFAT activation, which facilitates the interaction between CNS2 and Foxp3 promoter. Thus, epigenetically marked cis-elements can protect cell identity by sensing key environmental cues central to both cell identity formation and functional plasticity without interfering with initial cell differentiation.
Copyright © 2014 Elsevier Inc. All rights reserved.
Figures



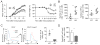


Comment in
-
Treg identity protection by an epigenetic switch.Cell Cycle. 2014;13(20):3159-60. doi: 10.4161/15384101.2014.969996. Cell Cycle. 2014. PMID: 25485491 Free PMC article. No abstract available.
References
-
- Burchill MA, Yang J, Vogtenhuber C, Blazar BR, Farrar MA. IL-2 receptor beta-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J Immunol. 2007;178:280–290. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

