The X gene of adeno-associated virus 2 (AAV2) is involved in viral DNA replication
- PMID: 25127256
- PMCID: PMC4134208
- DOI: 10.1371/journal.pone.0104596
The X gene of adeno-associated virus 2 (AAV2) is involved in viral DNA replication
Abstract
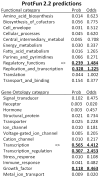
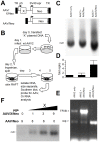
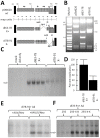
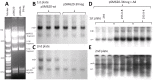
Adeno-associated virus (AAV) (type 2) is a popular human gene therapy vector with a long active transgene expression period and no reported vector-induced adverse reactions. Yet the basic molecular biology of this virus has not been fully addressed. One potential gene at the far 3' end of the AAV2 genome, previously referred to as X (nt 3929 to 4393), overlapping the 3' end of the cap gene, has never been characterized, although we did previously identify a promoter just up-stream (p81). Computer analysis suggested that X was involved in replication and transcription. The X protein was identified during active AAV2 replication using a polyclonal antibody against a peptide starting at amino acid 98. Reagents for the study of X included an AAV2 deletion mutant (dl78-91), a triple nucleotide substitution mutant that destroys all three 5' AUG-initiation products of X, with no effect on the cap coding sequence, and X-positive-293 cell lines. Here, we found that X up-regulated AAV2 DNA replication in differentiating keratinocytes (without helper virus, autonomous replication) and in various forms of 293 cell-based assays with help from wild type adenovirus type 5 (wt Ad5) or Ad5 helper plasmid (pHelper). The strongest contribution by X was seen in increasing wt AAV2 DNA replication in keratinocytes and dl78-91 in Ad5-infected X-positive-293 cell lines (both having multi-fold effects). Mutating the X gene in pAAV-RC (pAAV-RC-3Xneg) yielded approximately a ∼33% reduction in recombinant AAV vector DNA replication and virion production, but a larger effect was seen when using this same X-knockout AAV helper plasmid in X-positive-293 cell lines versus normal 293 cells (again, multi-fold). Taken together these data strongly suggest that AAV2 X encodes a protein involved in the AAV life cycle, particularly in increasing AAV2 DNA replication, and suggests that further studies are warranted.
Conflict of interest statement
Figures




References
-
- Hermonat PL (1984) Genetic analysis and utilization of adeno-associated virus as a mammalian cloning vector. Univ FL Dissertation, Available: http://archive.org/details/geneticanalysisu00herm.
-
- Hermonat PL (2014) The first adeno-associated virus gene transfer experiment, 1983. In press Human Gene Therapy - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

