CDK9-mediated transcription elongation is required for MYC addiction in hepatocellular carcinoma
- PMID: 25128497
- PMCID: PMC4197965
- DOI: 10.1101/gad.244368.114
CDK9-mediated transcription elongation is required for MYC addiction in hepatocellular carcinoma
Abstract
One-year survival rates for newly diagnosed hepatocellular carcinoma (HCC) are <50%, and unresectable HCC carries a dismal prognosis owing to its aggressiveness and the undruggable nature of its main genetic drivers. By screening a custom library of shRNAs directed toward known drug targets in a genetically defined Myc-driven HCC model, we identified cyclin-dependent kinase 9 (Cdk9) as required for disease maintenance. Pharmacological or shRNA-mediated CDK9 inhibition led to robust anti-tumor effects that correlated with MYC expression levels and depended on the role that both CDK9 and MYC exert in transcription elongation. Our results establish CDK9 inhibition as a therapeutic strategy for MYC-overexpressing liver tumors and highlight the relevance of transcription elongation in the addiction of cancer cells to MYC.
Keywords: CDK9; MYC; RNAi screen; oncogene addiction; transcription elongation.
© 2014 Huang et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
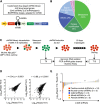
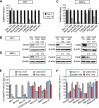
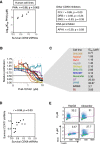

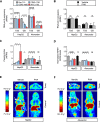
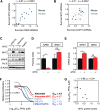
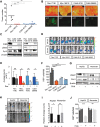
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
