Turning the spotlight on protein-lipid interactions in cells
- PMID: 25129056
- PMCID: PMC4206213
- DOI: 10.1016/j.cbpa.2014.07.015
Turning the spotlight on protein-lipid interactions in cells
Abstract
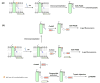
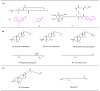
Protein function is largely dependent on coordinated and dynamic interactions of the protein with biomolecules including other proteins, nucleic acids and lipids. Although powerful methods for global profiling of protein-protein and protein-nucleic acid interactions are available, proteome-wide mapping of protein-lipid interactions is still challenging and rarely performed. The emergence of bifunctional lipid probes with photoactivatable and clickable groups offers new chemical tools for globally profiling protein-lipid interactions under cellular contexts. In this review, we summarize recent advances in the development of bifunctional lipid probes for studying protein-lipid interactions. We also highlight how in vivo photocrosslinking reactions contribute to the characterization of lipid-binding proteins and lipidation-mediated protein-protein interactions.
Copyright © 2014. Published by Elsevier Ltd.
Figures

Similar articles
-
In Vitro Approaches for Detection of Protein-Lipid Interactions and Protein-to-Protein Transport of Lipids Using Bifunctional Lipid Analogs.Methods Mol Biol. 2025;2888:249-258. doi: 10.1007/978-1-0716-4318-1_17. Methods Mol Biol. 2025. PMID: 39699736
-
Fat & fabulous: bifunctional lipids in the spotlight.Biochim Biophys Acta. 2014 Aug;1841(8):1022-30. doi: 10.1016/j.bbalip.2014.01.003. Epub 2014 Jan 15. Biochim Biophys Acta. 2014. PMID: 24440797 Review.
-
Recent developments and applications of clickable photoprobes in medicinal chemistry and chemical biology.Future Med Chem. 2015;7(16):2143-71. doi: 10.4155/fmc.15.136. Epub 2015 Oct 29. Future Med Chem. 2015. PMID: 26511756 Review.
-
Bifunctional Lipid-Derived Affinity-Based Probes (AfBPs) for Analysis of Lipid-Protein Interactome.Acc Chem Res. 2022 Dec 20;55(24):3663-3674. doi: 10.1021/acs.accounts.2c00593. Epub 2022 Dec 9. Acc Chem Res. 2022. PMID: 36484537
-
Global profiling of protein lipidation using chemical proteomic technologies.Curr Opin Chem Biol. 2015 Feb;24:48-57. doi: 10.1016/j.cbpa.2014.10.016. Epub 2014 Nov 15. Curr Opin Chem Biol. 2015. PMID: 25461723 Free PMC article. Review.
Cited by
-
Photoaffinity labelling strategies for mapping the small molecule-protein interactome.Org Biomol Chem. 2021 Sep 22;19(36):7792-7809. doi: 10.1039/d1ob01353j. Org Biomol Chem. 2021. PMID: 34549230 Free PMC article. Review.
-
Druggable Lipid Binding Sites in Pentameric Ligand-Gated Ion Channels and Transient Receptor Potential Channels.Front Physiol. 2022 Jan 4;12:798102. doi: 10.3389/fphys.2021.798102. eCollection 2021. Front Physiol. 2022. PMID: 35069257 Free PMC article. Review.
-
YPIBP: A repository for phosphoinositide-binding proteins in yeast.Comput Struct Biotechnol J. 2021 Jun 24;19:3692-3707. doi: 10.1016/j.csbj.2021.06.035. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 34285772 Free PMC article.
-
Emerging Diversity in Lipid-Protein Interactions.Chem Rev. 2019 May 8;119(9):5775-5848. doi: 10.1021/acs.chemrev.8b00451. Epub 2019 Feb 13. Chem Rev. 2019. PMID: 30758191 Free PMC article. Review.
-
Photoactivatable Glycolipid Probes for Identifying Mycolate-Protein Interactions in Live Mycobacteria.J Am Chem Soc. 2020 Apr 29;142(17):7725-7731. doi: 10.1021/jacs.0c01065. Epub 2020 Apr 20. J Am Chem Soc. 2020. PMID: 32293873 Free PMC article.
References
-
- van Meer G, de Kroon AIPM. Lipid map of the mammalian cell. J Cell Sci. 2011;124:5–8. - PubMed
-
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. - PubMed
-
- Wymann MP, Schneiter R. Lipid signalling in disease. Nat Rev Mol Cell Biol. 2008;9:162–176. - PubMed
-
- Santos CR, Schulze A. Lipid metabolism in cancer. FEBS J. 2012;279:2610–2623. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

