HIV-1 envelope protein gp41: an NMR study of dodecyl phosphocholine embedded gp41 reveals a dynamic prefusion intermediate conformation
- PMID: 25132083
- PMCID: PMC4156547
- DOI: 10.1016/j.str.2014.06.016
HIV-1 envelope protein gp41: an NMR study of dodecyl phosphocholine embedded gp41 reveals a dynamic prefusion intermediate conformation
Abstract
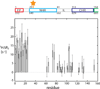
Human immunodeficiency viral (HIV-1) fusion is mediated by the viral envelope gp120/gp41 complex (ENVelope glycoprotein). After gp120 shedding, gp41 is exposed and elicits membrane fusion via a cascade of conformational changes. In contrast to prefusion and postfusion conformation, little is known about any intermediate conformation. We report on a solution NMR investigation of homotrimeric HIV-1 gp41(27-194), comprising the transmembrane region and reconstituted in dodecyl phosphocholine (DPC) micelles. The protein is mainly α-helical, but experiences internal dynamics on the nanosecond and micro to millisecond time scale and transient α-helical behavior for certain residues in the N-terminal heptad repeat (NHR). Strong lipid interactions are observed, in particular for C-terminal residues of the NHR and imunodominant loop region connecting NHR and C-terminal heptad repeat (CHR). Our data indicate an extended conformation with features anticipated for a prefusion intermediate, presumably in exchange with a lowly populated postfusion six-helical bundle conformation.
Copyright © 2014 Elsevier Ltd. All rights reserved.
Figures





Comment in
-
Capturing glimpses of an elusive HIV gp41 prehairpin fusion intermediate.Structure. 2014 Sep 2;22(9):1225-1226. doi: 10.1016/j.str.2014.08.010. Structure. 2014. PMID: 25185826 Free PMC article.
References
-
- Chan DC, Fass D, Berger JM, Kim PS. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

