The cistrome and gene signature of androgen receptor splice variants in castration resistant prostate cancer cells
- PMID: 25132238
- PMCID: PMC4411637
- DOI: 10.1016/j.juro.2014.08.043
The cistrome and gene signature of androgen receptor splice variants in castration resistant prostate cancer cells
Abstract
Purpose: Spliced variant forms of androgen receptor were recently identified in castration resistant prostate cancer cell lines and clinical samples. We identified the cistrome and gene signature of androgen receptor splice variants in castration resistant prostate cancer cell lines and determined the clinical significance of androgen receptor splice variant regulated genes.
Materials and methods: The castration resistant prostate cancer cell line 22Rv1, which expresses full-length androgen receptor and androgen receptor splice variants endogenously, was used as the research model. We established 22Rv1-ARFL(-)/ARV(+) and 22Rv1-ARFL(-)/ARV(-) through RNA interference. Chromatin immunoprecipitation coupled with next generation sequencing and microarray techniques were used to identify the cistrome and gene expression profiles of androgen receptor splice variants in the absence of androgen.
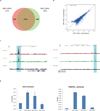
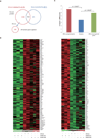
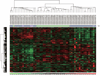
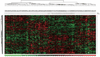
Results: Androgen receptor splice variant binding sites were identified in 22Rv1-ARFL(-)/ARV(+). A gene set was regulated uniquely by androgen receptor splice variants but not by full-length androgen receptor in the absence of androgen. Integrated analysis revealed that some genes were directly modulated by androgen receptor splice variants. Unsupervised clustering analysis showed that the androgen receptor splice variant gene signature differentiated benign from malignant prostate tissue as well as localized prostate cancer from metastatic castration resistant prostate cancer specimens. Some genes that were modulated uniquely by androgen receptor splice variants also correlated with histological grade and biochemical failure.
Conclusions: Androgen receptor splice variants can bind to DNA independent of full-length androgen receptor in the absence of androgen and modulate a unique set of genes that is not regulated by full-length androgen receptor. The androgen receptor splice variant gene signature correlates with disease progression. It distinguishes primary cancer from castration resistant prostate cancer specimens and benign from malignant prostate specimens.
Keywords: alternative splicing; androgen; castration-resistant; disease progression; prostatic neoplasms; receptors; transcriptome.
Copyright © 2015 American Urological Association Education and Research, Inc. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures
References
-
- Siegel R, Ma J, Zou Z, et al. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9. - PubMed
-
- Debes JD, Tindall DJ. Mechanisms of androgen-refractory prostate cancer. N Engl J Med. 2004;351:1488. - PubMed
-
- Lu J, Tindall DJ. The role of androgen receptor-regulated genes in prostate cancer progression. In: Socorro S, editor. Androgen receptors : structural biology, genetics and molecular defects. United States: NOVA SCIENCE PUBLISHERS; 2014. pp. 107–128.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

