Leaf development: a cellular perspective
- PMID: 25132838
- PMCID: PMC4116805
- DOI: 10.3389/fpls.2014.00362
Leaf development: a cellular perspective
Abstract
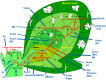
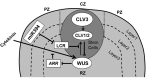
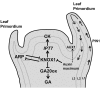
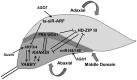
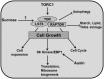
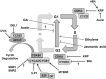
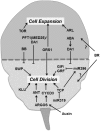
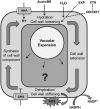
Through its photosynthetic capacity the leaf provides the basis for growth of the whole plant. In order to improve crops for higher productivity and resistance for future climate scenarios, it is important to obtain a mechanistic understanding of leaf growth and development and the effect of genetic and environmental factors on the process. Cells are both the basic building blocks of the leaf and the regulatory units that integrate genetic and environmental information into the developmental program. Therefore, to fundamentally understand leaf development, one needs to be able to reconstruct the developmental pathway of individual cells (and their progeny) from the stem cell niche to their final position in the mature leaf. To build the basis for such understanding, we review current knowledge on the spatial and temporal regulation mechanisms operating on cells, contributing to the formation of a leaf. We focus on the molecular networks that control exit from stem cell fate, leaf initiation, polarity, cytoplasmic growth, cell division, endoreduplication, transition between division and expansion, expansion and differentiation and their regulation by intercellular signaling molecules, including plant hormones, sugars, peptides, proteins, and microRNAs. We discuss to what extent the knowledge available in the literature is suitable to be applied in systems biology approaches to model the process of leaf growth, in order to better understand and predict leaf growth starting with the model species Arabidopsis thaliana.
Keywords: developmental pathway; leaf growth; modeling; plant hormones; stress.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

