The bacterial translation stress response
- PMID: 25135187
- PMCID: PMC4227928
- DOI: 10.1111/1574-6976.12083
The bacterial translation stress response
Abstract
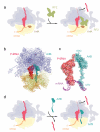
Throughout their life, bacteria need to sense and respond to environmental stress. Thus, such stress responses can require dramatic cellular reprogramming, both at the transcriptional as well as the translational level. This review focuses on the protein factors that interact with the bacterial translational apparatus to respond to and cope with different types of environmental stress. For example, the stringent factor RelA interacts with the ribosome to generate ppGpp under nutrient deprivation, whereas a variety of factors have been identified that bind to the ribosome under unfavorable growth conditions to shut-down (RelE, pY, RMF, HPF and EttA) or re-program (MazF, EF4 and BipA) translation. Additional factors have been identified that rescue ribosomes stalled due to stress-induced mRNA truncation (tmRNA, ArfA, ArfB), translation of unfavorable protein sequences (EF-P), heat shock-induced subunit dissociation (Hsp15), or antibiotic inhibition (TetM, FusB). Understanding the mechanism of how the bacterial cell responds to stress will not only provide fundamental insight into translation regulation, but will also be an important step to identifying new targets for the development of novel antimicrobial agents.
Keywords: antibiotic stress; mRNA truncation; nutrient depletion; stationary phase; toxin-antitoxin modules; translational stalling.
© 2014 Federation of European Microbiological Societies. Published by John Wiley & Sons Ltd. All rights reserved.
Figures

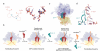


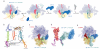




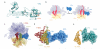
References
-
- Abe T, Sakaki K, Fujihara A, Ujiie H, Ushida C, Himeno H, Sato T, Muto A. tmRNA-dependent trans-translation is required for sporulation in Bacillus subtilis. Molecular microbiology. 2008;69:1491–1498. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

