cPKC regulates interphase nuclear size during Xenopus development
- PMID: 25135933
- PMCID: PMC4137061
- DOI: 10.1083/jcb.201406004
cPKC regulates interphase nuclear size during Xenopus development
Abstract
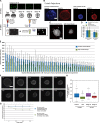
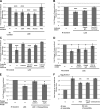
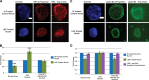
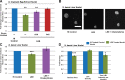
Dramatic changes in cell and nuclear size occur during development and differentiation, and aberrant nuclear size is associated with many disease states. However, the mechanisms that regulate nuclear size are largely unknown. A robust system for investigating nuclear size is early Xenopus laevis development, during which reductions in nuclear size occur without changes in DNA content. To identify cellular factors that regulate nuclear size during development, we developed a novel nuclear resizing assay wherein nuclei assembled in Xenopus egg extract become smaller in the presence of cytoplasmic interphase extract isolated from post-gastrula Xenopus embryos. We show that nuclear shrinkage depends on conventional protein kinase C (cPKC). Increased nuclear cPKC localization and activity and decreased nuclear association of lamins mediate nuclear size reductions during development, and manipulating cPKC activity in vivo during interphase alters nuclear size in the embryo. We propose a model of steady-state nuclear size regulation whereby nuclear expansion is balanced by an active cPKC-dependent mechanism that reduces nuclear size.
© 2014 Edens and Levy.
Figures
Comment in
-
Development: Honey, I shrunk the nucleus.Nat Rev Mol Cell Biol. 2014 Oct;15(10):633. doi: 10.1038/nrm3872. Epub 2014 Sep 4. Nat Rev Mol Cell Biol. 2014. PMID: 25186132 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

