ExaBayes: massively parallel bayesian tree inference for the whole-genome era
- PMID: 25135941
- PMCID: PMC4166930
- DOI: 10.1093/molbev/msu236
ExaBayes: massively parallel bayesian tree inference for the whole-genome era
Abstract
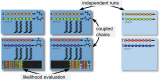
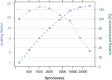
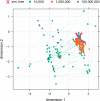
Modern sequencing technology now allows biologists to collect the entirety of molecular evidence for reconstructing evolutionary trees. We introduce a novel, user-friendly software package engineered for conducting state-of-the-art Bayesian tree inferences on data sets of arbitrary size. Our software introduces a nonblocking parallelization of Metropolis-coupled chains, modifications for efficient analyses of data sets comprising thousands of partitions and memory saving techniques. We report on first experiences with Bayesian inferences at the whole-genome level using the SuperMUC supercomputer and simulated data.
Keywords: Bayesian statistics; parallelization; phylogenetic inference; software; whole-genome analyses.
© The Author 2014. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures
References
-
- Altekar G, Dwarkadas S, Huelsenbeck JP, Ronquist F. Parallel Metropolis coupled Markov chain Monte Carlo for Bayesian phylogenetic inference. Bioinformatics. 2004;20(3):407–415. - PubMed
-
- Dunn CW, Hejnol A, Matus DQ, Pang K, Browne WE, Smith SA, Seaver E, Rouse GW, Obst M, Edgecombe GD, et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452(7188):745–749. - PubMed
-
- Fitch WM, Margoliash E. Construction of phylogenetic trees. Science. 1967;155(760):279–284. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

