DNA modifications in the mammalian brain
- PMID: 25135973
- PMCID: PMC4142033
- DOI: 10.1098/rstb.2013.0512
DNA modifications in the mammalian brain
Abstract
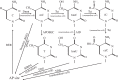
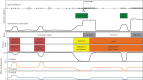
DNA methylation is a crucial epigenetic mark in mammalian development, genomic imprinting, X-inactivation, chromosomal stability and suppressing parasitic DNA elements. DNA methylation in neurons has also been suggested to play important roles for mammalian neuronal functions, and learning and memory. In this review, we first summarize recent discoveries and fundamental principles of DNA modifications in the general epigenetics field. We then describe the profiles of different DNA modifications in the mammalian brain genome. Finally, we discuss roles of DNA modifications in mammalian brain development and function.
Keywords: 5-hydroxymethylcytosine; 5-methylcytosine; DNA methylation; learning and memory.
© 2014 The Author(s) Published by the Royal Society. All rights reserved.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
