Regulation of histone H3K4 methylation in brain development and disease
- PMID: 25135975
- PMCID: PMC4142035
- DOI: 10.1098/rstb.2013.0514
Regulation of histone H3K4 methylation in brain development and disease
Abstract
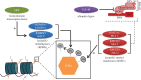
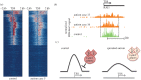
The growing list of mutations implicated in monogenic disorders of the developing brain includes at least seven genes (ARX, CUL4B, KDM5A, KDM5C, KMT2A, KMT2C, KMT2D) with loss-of-function mutations affecting proper regulation of histone H3 lysine 4 methylation, a chromatin mark which on a genome-wide scale is broadly associated with active gene expression, with its mono-, di- and trimethylated forms differentially enriched at promoter and enhancer and other regulatory sequences. In addition to these rare genetic syndromes, dysregulated H3K4 methylation could also play a role in the pathophysiology of some cases diagnosed with autism or schizophrenia, two conditions which on a genome-wide scale are associated with H3K4 methylation changes at hundreds of loci in a subject-specific manner. Importantly, the reported alterations for some of the diseased brain specimens included a widespread broadening of H3K4 methylation profiles at gene promoters, a process that could be regulated by the UpSET(KMT2E/MLL5)-histone deacetylase complex. Furthermore, preclinical studies identified maternal immune activation, parental care and monoaminergic drugs as environmental determinants for brain-specific H3K4 methylation. These novel insights into the epigenetic risk architectures of neurodevelopmental disease will be highly relevant for efforts aimed at improved prevention and treatment of autism and psychosis spectrum disorders.
Keywords: autism; chromatin; epigenetic; histone; nucleosome; schizophrenia.
© 2014 The Author(s) Published by the Royal Society. All rights reserved.
Figures
References
-
- Siegmund KD, Connor CM, Campan M, Long TI, Weisenberger DJ, Biniszkiewicz D, Jaenisch R, Laird PW, Akbarian S. 2007. DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PLoS ONE 2, e895 (10.1371/journal.pone.0000895) - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
