Adaptive, convergent origins of the pygmy phenotype in African rainforest hunter-gatherers
- PMID: 25136101
- PMCID: PMC4156716
- DOI: 10.1073/pnas.1402875111
Adaptive, convergent origins of the pygmy phenotype in African rainforest hunter-gatherers
Abstract
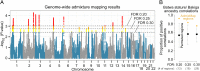
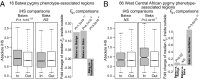
The evolutionary history of the human pygmy phenotype (small body size), a characteristic of African and Southeast Asian rainforest hunter-gatherers, is largely unknown. Here we use a genome-wide admixture mapping analysis to identify 16 genomic regions that are significantly associated with the pygmy phenotype in the Batwa, a rainforest hunter-gatherer population from Uganda (east central Africa). The identified genomic regions have multiple attributes that provide supporting evidence of genuine association with the pygmy phenotype, including enrichments for SNPs previously associated with stature variation in Europeans and for genes with growth hormone receptor and regulation functions. To test adaptive evolutionary hypotheses, we computed the haplotype-based integrated haplotype score (iHS) statistic and the level of population differentiation (FST) between the Batwa and their agricultural neighbors, the Bakiga, for each genomic SNP. Both |iHS| and FST values were significantly higher for SNPs within the Batwa pygmy phenotype-associated regions than the remainder of the genome, a signature of polygenic adaptation. In contrast, when we expanded our analysis to include Baka rainforest hunter-gatherers from Cameroon and Gabon (west central Africa) and Nzebi and Nzime neighboring agriculturalists, we did not observe elevated |iHS| or FST values in these genomic regions. Together, these results suggest adaptive and at least partially convergent origins of the pygmy phenotype even within Africa, supporting the hypothesis that small body size confers a selective advantage for tropical rainforest hunter-gatherers but raising questions about the antiquity of this behavior.
Keywords: convergent evolution; human evolutionary ecology; human hunter-gatherers; population genomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Perry GH, Dominy NJ. Evolution of the human pygmy phenotype. Trends Ecol Evol. 2009;24(4):218–225. - PubMed
-
- Migliano AB, et al. Evolution of the pygmy phenotype: Evidence of positive selection fro genome-wide scans in African, Asian, and Melanesian pygmies. Hum Biol. 2013;85(1-3):251–284. - PubMed
-
- Cavalli-Sforza LL. African pygmies: An evaluation of the state of research. In: Cavalli-Sforza LL, editor. African Pygmies. Orlando, FL: Academic Press; 1986. pp. 361–426.
-
- Diamond JM. Anthropology. Why are pygmies small? Nature. 1991;354(6349):111–112. - PubMed
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

