Drug Resistance Mutations Alter Dynamics of Inhibitor-Bound HIV-1 Protease
- PMID: 25136270
- PMCID: PMC4132871
- DOI: 10.1021/ct4010454
Drug Resistance Mutations Alter Dynamics of Inhibitor-Bound HIV-1 Protease
Abstract
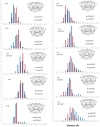
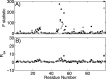
Under the selective pressure of therapy, HIV-1 protease mutants resistant to inhibitors evolve to confer drug resistance. Such mutations can impact both the dynamics and structures of the bound and unbound forms of the enzyme. Flap+ is a multidrug-resistant variant of HIV-1 protease with a combination of primary and secondary resistance mutations (L10I, G48V, I54V, V82A) and a strikingly altered thermodynamic profile for darunavir (DRV) binding relative to the wild-type protease. We elucidated the impact of these mutations on protein dynamics in the DRV-bound state using molecular dynamics simulations and NMR relaxation experiments. Both methods concur in that the conformational ensemble and dynamics of protease are impacted by the drug resistance mutations in Flap+ variant. Surprisingly this change in ensemble dynamics is different from that observed in the unliganded form of the same variant (Cai, Y. et al. J. Chem. Theory Comput.2012, 8, 3452-3462). Our comparative analysis of both inhibitor-free and bound states presents a comprehensive picture of the altered dynamics in drug-resistant mutant HIV-1 protease and underlies the importance of incorporating dynamic analysis of the whole system, including the unliganded state, into revealing drug resistance mechanisms.
Figures





References
-
- Altman M. D.; Ali A.; Reddy G. S.; Nalam M. N.; Anjum S. G.; Cao H.; Chellappan S.; Kairys V.; Fernandes M. X.; Gilson M. K.; Schiffer C. A.; Rana T. M.; Tidor B. HIV-1 protease inhibitors from inverse design in the substrate envelope exhibit subnanomolar binding to drug-resistant variants. J. Am. Chem. Soc. 2008, 130, 6099–113. - PMC - PubMed
- Nalam M. N.; Ali A.; Reddy G. S.; Cao H.; Anjum S. G.; Altman M. D.; Yilmaz N. K.; Tidor B.; Rana T. M.; Schiffer C. A. Substrate Envelope-Designed Potent HIV-1 Protease Inhibitors to Avoid Drug Resistance. Chem. Biol. 2013, 20, 1116–24. - PMC - PubMed
- Ozen A.; Haliloglu T.; Schiffer C. A. Dynamics of Preferential Substrate Recognition in HIV-1 Protease: Redefining the Substrate Envelope. J. Mol. Biol. 2011, 410, 726–744. - PMC - PubMed
- Prabu-Jeyabalan M.; King N. M.; Nalivaika E. A.; Heilek-Snyder G.; Cammack N.; Schiffer C. A. Substrate envelope and drug resistance: crystal structure of RO1 in complex with wild-type human immunodeficiency virus type 1 protease. Antimicrob. Agents Chemother. 2006, 50, 1518–21. - PMC - PubMed
-
- Clemente J. C.; Hemrajani R.; Blum L. E.; Goodenow M. M.; Dunn B. M. Secondary mutations M36I and A71V in the human immunodeficiency virus type 1 protease can provide an advantage for the emergence of the primary mutation D30N. Biochemistry (Moscow) 2003, 42, 15029–35. - PubMed
- Clemente J. C.; Moose R. E.; Hemrajani R.; Whitford L. R.; Govindasamy L.; Reutzel R.; McKenna R.; Agbandje-McKenna M.; Goodenow M. M.; Dunn B. M. Comparing the accumulation of active- and nonactive-site mutations in the HIV-1 protease. Biochemistry (Moscow) 2004, 43, 12141–51. - PubMed
-
- Foulkes-Murzycki J. E.; Scott W. R.; Schiffer C. A. Hydrophobic sliding: a possible mechanism for drug resistance in human immunodeficiency virus type 1 protease. Structure 2007, 15, 225–33. - PMC - PubMed
- Mittal S.; Cai Y.; Nalam M. N.; Bolon D. N.; Schiffer C. A. Hydrophobic core flexibility modulates enzyme activity in HIV-1 protease. J. Am. Chem. Soc. 2012, 134, 4163–8. - PMC - PubMed
-
- Freedberg D. I.; Ishima R.; Jacob J.; Wang Y. X.; Kustanovich I.; Louis J. M.; Torchia D. A. Rapid structural fluctuations of the free HIV protease flaps in solution: relationship to crystal structures and comparison with predictions of dynamics calculations. Protein Sci. 2002, 11, 221–32. - PMC - PubMed
- Galiano L.; Bonora M.; Fanucci G. E. Interflap distances in HIV-1 protease determined by pulsed EPR measurements. J. Am. Chem. Soc. 2007, 129, 11004–5. - PubMed
- Hornak V.; Okur A.; Rizzo R. C.; Simmerling C. HIV-1 protease flaps spontaneously close to the correct structure in simulations following manual placement of an inhibitor into the open state. J. Am. Chem. Soc. 2006, 128, 2812–3. - PMC - PubMed
- Ishima R.; Freedberg D. I.; Wang Y. X.; Louis J. M.; Torchia D. A. Flap opening and dimer-interface flexibility in the free and inhibitor-bound HIV protease, and their implications for function. Structure 1999, 7, 1047–55. - PubMed
- Ishima R.; Louis J. M. A diverse view of protein dynamics from NMR studies of HIV-1 protease flaps. Proteins 2008, 70, 1408–15. - PubMed
- Louis J. M.; Ishima R.; Nesheiwat I.; Pannell L. K.; Lynch S. M.; Torchia D. A.; Gronenborn A. M. Revisiting monomeric HIV-1 protease. Characterization and redesign for improved properties. J. Biol. Chem. 2003, 278, 6085–92. - PubMed
- Perryman A. L.; Lin J. H.; McCammon J. A. HIV-1 protease molecular dynamics of a wild-type and of the V82F/I84V mutant: possible contributions to drug resistance and a potential new target site for drugs. Protein Sci. 2004, 13, 1108–23. - PMC - PubMed
- Scott W. R.; Schiffer C. A. Curling of flap tips in HIV-1 protease as a mechanism for substrate entry and tolerance of drug resistance. Structure 2000, 8, 1259–65. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
