Divided we stand: splitting synthetic cells for their proliferation
- PMID: 25136387
- PMCID: PMC4127174
- DOI: 10.1007/s11693-014-9145-7
Divided we stand: splitting synthetic cells for their proliferation
Abstract
With the recent dawn of synthetic biology, the old idea of man-made artificial life has gained renewed interest. In the context of a bottom-up approach, this entails the de novo construction of synthetic cells that can autonomously sustain themselves and proliferate. Reproduction of a synthetic cell involves the synthesis of its inner content, replication of its information module, and growth and division of its shell. Theoretical and experimental analysis of natural cells shows that, whereas the core synthesis machinery of the information module is highly conserved, a wide range of solutions have been realized in order to accomplish division. It is therefore to be expected that there are multiple ways to engineer division of synthetic cells. Here we survey the field and review potential routes that can be explored to accomplish the division of bottom-up designed synthetic cells. We cover a range of complexities from simple abiotic mechanisms involving splitting of lipid-membrane-encapsulated vesicles due to physical or chemical principles, to potential division mechanisms of synthetic cells that are based on prokaryotic division machineries.
Keywords: Cell division; Minimal cells; Synthetic cells; Vesicle splitting.
Figures
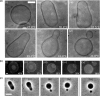


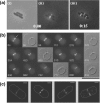


References
-
- Aarsman MEG, Piette A, Fraipont C, Vinkenvleugel TMF, Nguyen-Distèche M, den Blaauwen T. Maturation of the Escherichia coli divisome occurs in two steps. Mol Microbiol. 2005;55:1631–1645. - PubMed
-
- Adams DW, Errington J. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat Rev Microbiol. 2009;7:642–653. - PubMed
-
- Akerlund T, Gullbrand B, Nordström K. Effects of the Min system on nucleoid segregation in Escherichia coli. Microbiology. 2002;148:3213–3222. - PubMed
-
- Albers SV, Meyer BH. The archaeal cell envelope. Nat Rev Microbiol. 2011;9:414–426. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

