Refined candidate region for F4ab/ac enterotoxigenic Escherichia coli susceptibility situated proximal to MUC13 in pigs
- PMID: 25137053
- PMCID: PMC4138166
- DOI: 10.1371/journal.pone.0105013
Refined candidate region for F4ab/ac enterotoxigenic Escherichia coli susceptibility situated proximal to MUC13 in pigs
Abstract
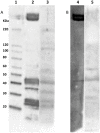
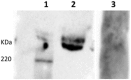
F4 enterotoxigenic Escherichia coli (F4 ETEC) are an important cause of diarrhea in neonatal and newly-weaned pigs. Based on the predicted differential O-glycosylation patterns of the 2 MUC13 variants (MUC13A and MUC13B) in F4ac ETEC susceptible and F4ac ETEC resistant pigs, the MUC13 gene was recently proposed as the causal gene for F4ac ETEC susceptibility. Because the absence of MUC13 on Western blot from brush border membrane vesicles of F4ab/acR+ pigs and the absence of F4ac attachment to immunoprecipitated MUC13 could not support this hypothesis, a new GWAS study was performed using 52 non-adhesive and 68 strong adhesive pigs for F4ab/ac ETEC originating from 5 Belgian farms. A refined candidate region (chr13: 144,810,100-144,993,222) for F4ab/ac ETEC susceptibility was identified with MUC13 adjacent to the distal part of the region. This candidate region lacks annotated genes and contains a sequence gap based on the sequence of the porcine GenomeBuild 10.2. We hypothesize that a porcine orphan gene or trans-acting element present in the identified candidate region has an effect on the glycosylation of F4 binding proteins and therefore determines the F4ab/ac ETEC susceptibility in pigs.
Conflict of interest statement
Figures



References
-
- Gibbons RA, Sellwood R, Burrows M, Hunter PA (1977) Inheritance of resistance to neonatal E. coli diarrhea in the pig: examination of the genetic system. Theor Appl Genet 51: 65–70. - PubMed
-
- Fairbrother JM, Nadeau E, Gyles CL (2005) Escherichia coli in postweaning diarrhea in pigs: an update on bacterial types, pathogenesis, and prevention strategies. Anim Health Res Rev 6: 17–39. - PubMed
-
- Zhang W, Zhao M, Ruesch L, Omot A, Francis D (2007) Prevalence of virulence genes in Escherichia coli strains recently isolated from young pigs with diarrhea in the US. Vet Microbiol 123: 145–152. - PubMed
-
- Wang J, Jiang SW, Chen XH, Liu ZL, Peng J (2006) Prevalence of fimbrial antigen (K88 variants, K99 and 987P) of enterotoxigenic Escherichia coli from neonatal and postweaning piglets with diarrhea in central China. Asian-Aust J Anim Sci 19: 1342–1346.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

