Catalytic profile of Arabidopsis peroxidases, AtPrx-2, 25 and 71, contributing to stem lignification
- PMID: 25137070
- PMCID: PMC4138150
- DOI: 10.1371/journal.pone.0105332
Catalytic profile of Arabidopsis peroxidases, AtPrx-2, 25 and 71, contributing to stem lignification
Abstract
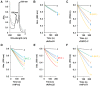
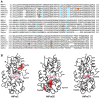
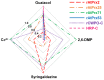
Lignins are aromatic heteropolymers that arise from oxidative coupling of lignin precursors, including lignin monomers (p-coumaryl, coniferyl, and sinapyl alcohols), oligomers, and polymers. Whereas plant peroxidases have been shown to catalyze oxidative coupling of monolignols, the oxidation activity of well-studied plant peroxidases, such as horseradish peroxidase C (HRP-C) and AtPrx53, are quite low for sinapyl alcohol. This characteristic difference has led to controversy regarding the oxidation mechanism of sinapyl alcohol and lignin oligomers and polymers by plant peroxidases. The present study explored the oxidation activities of three plant peroxidases, AtPrx2, AtPrx25, and AtPrx71, which have been already shown to be involved in lignification in the Arabidopsis stem. Recombinant proteins of these peroxidases (rAtPrxs) were produced in Escherichia coli as inclusion bodies and successfully refolded to yield their active forms. rAtPrx2, rAtPrx25, and rAtPrx71 were found to oxidize two syringyl compounds (2,6-dimethoxyphenol and syringaldazine), which were employed here as model monolignol compounds, with higher specific activities than HRP-C and rAtPrx53. Interestingly, rAtPrx2 and rAtPrx71 oxidized syringyl compounds more efficiently than guaiacol. Moreover, assays with ferrocytochrome c as a substrate showed that AtPrx2, AtPrx25, and AtPrx71 possessed the ability to oxidize large molecules. This characteristic may originate in a protein radical. These results suggest that the plant peroxidases responsible for lignin polymerization are able to directly oxidize all lignin precursors.
Conflict of interest statement
Figures

References
-
- Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546. - PubMed
-
- Sarkanen KV (1971) Precursors and their polymerization. In: Sarkanen KV and Ludwig CH (eds) Lignins, Occurrence, Formation, Structure and Reactions. Wiley-Interscience, New York, pp.95–163.
-
- Almagro L, Gómez Ros LV, Belchi-Navarro S, Bru R, Ros Barceló A, et al. (2009) Class III peroxidases in plant defence reactions. J Exp Bot 60: 377–390. - PubMed
-
- El Mansouri I, Mercado JA, Santiago-Dómenech N, Pliego-Alfaro F, Valpuesta V, et al. (1999) Biochemical and phenotypical characterization of transgenic tomato plants overexpressing a basic peroxidase. Physiol Plant 106: 355–362.
-
- Li Y, Kajita S, Kawai S, Katayama Y, Morohoshi N (2003) Down-regulation of an anionic peroxidase in transgenic aspen and its effect on lignin characteristics. J Plant Res 116: 175–182. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

