The MicroRNA Biology of the Mammalian Nucleus
- PMID: 25137140
- PMCID: PMC4221600
- DOI: 10.1038/mtna.2014.40
The MicroRNA Biology of the Mammalian Nucleus
Abstract
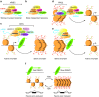
MicroRNAs (miRNAs) are a class of genome-encoded small RNAs that are primarily considered to be post-transcriptional negative regulators of gene expression acting in the cytoplasm. Over a decade of research has focused on this canonical paradigm of miRNA function, with many success stories. Indeed, miRNAs have been identified that act as master regulators of a myriad of cellular processes, and many miRNAs are promising therapeutic targets or disease biomarkers. However, it is becoming increasingly apparent that the canonical view of miRNA function is incomplete. Several lines of evidence now point to additional functions for miRNAs in the nucleus of the mammalian cell. The majority of cellular miRNAs are present in both the nucleus and the cytoplasm, and certain miRNAs show specific nuclear enrichment. Additionally, some miRNAs colocalize with sub-nuclear structures such as the nucleolus and chromatin. Multiple components of the miRNA processing machinery are present in the nuclear compartment and are shuttled back and forth across the nuclear envelope. In the nucleus, miRNAs act to regulate the stability of nuclear transcripts, induce epigenetic alterations that either silence or activate transcription at specific gene promoters, and modulate cotranscriptional alternative splicing events. Nuclear miRNA-directed gene regulation constitutes a departure from the prevailing view of miRNA function and as such, warrants detailed further investigation.
Figures

References
-
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. - PubMed
-
- Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. - PubMed
-
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. - PubMed
-
- Filipowicz W, Jaskiewicz L, Kolb FA, Pillai RS. Post-transcriptional gene silencing by siRNAs and miRNAs. Curr Opin Struct Biol. 2005;15:331–341. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

