The cytoplasmic capping complex assembles on adapter protein nck1 bound to the proline-rich C-terminus of Mammalian capping enzyme
- PMID: 25137142
- PMCID: PMC4138027
- DOI: 10.1371/journal.pbio.1001933
The cytoplasmic capping complex assembles on adapter protein nck1 bound to the proline-rich C-terminus of Mammalian capping enzyme
Abstract
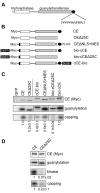
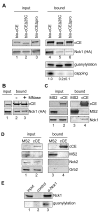
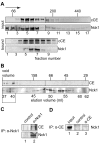
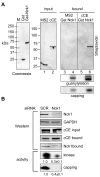
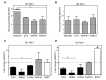
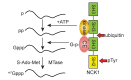
Cytoplasmic capping is catalyzed by a complex that contains capping enzyme (CE) and a kinase that converts RNA with a 5'-monophosphate end to a 5' diphosphate for subsequent addition of guanylic acid (GMP). We identify the proline-rich C-terminus as a new domain of CE that is required for its participation in cytoplasmic capping, and show the cytoplasmic capping complex assembles on Nck1, an adapter protein with functions in translation and tyrosine kinase signaling. Binding is specific to Nck1 and is independent of RNA. We show by sedimentation and gel filtration that Nck1 and CE are together in a larger complex, that the complex can assemble in vitro on recombinant Nck1, and Nck1 knockdown disrupts the integrity of the complex. CE and the 5' kinase are juxtaposed by binding to the adjacent domains of Nck1, and cap homeostasis is inhibited by Nck1 with inactivating mutations in each of these domains. These results identify a new domain of CE that is specific to its function in cytoplasmic capping, and a new role for Nck1 in regulating gene expression through its role as the scaffold for assembly of the cytoplasmic capping complex.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

