Regression of replication forks stalled by leading-strand template damage: I. Both RecG and RuvAB catalyze regression, but RuvC cleaves the holliday junctions formed by RecG preferentially
- PMID: 25138216
- PMCID: PMC4192490
- DOI: 10.1074/jbc.M114.587881
Regression of replication forks stalled by leading-strand template damage: I. Both RecG and RuvAB catalyze regression, but RuvC cleaves the holliday junctions formed by RecG preferentially
Abstract
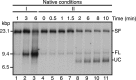
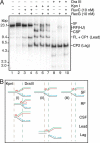
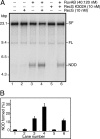
The orderly progression of replication forks formed at the origin of replication in Escherichia coli is challenged by encounters with template damage, slow moving RNA polymerases, and frozen DNA-protein complexes that stall the fork. These stalled forks are foci for genomic instability and must be reactivated. Many models of replication fork reactivation invoke nascent strand regression as an intermediate in the processing of the stalled fork. We have investigated the replication fork regression activity of RecG and RuvAB, two proteins commonly thought to be involved in the process, using a reconstituted DNA replication system where the replisome is stalled by collision with leading-strand template damage. We find that both RecG and RuvAB can regress the stalled fork in the presence of the replisome and SSB; however, RuvAB generates a completely unwound product consisting of the paired nascent leading and lagging strands, whereas RuvC cleaves the Holliday junction generated by RecG-catalyzed fork regression. We also find that RecG stimulates RuvAB-catalyzed regression, presumably because it is more efficient at generating the initial Holliday junction from the stalled fork.
Keywords: DNA Enzyme; DNA Recombination; DNA Repair; DNA Replication; Genomic Instability.
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures








Similar articles
-
The Biochemical Mechanism of Fork Regression in Prokaryotes and Eukaryotes-A Single Molecule Comparison.Int J Mol Sci. 2022 Aug 3;23(15):8613. doi: 10.3390/ijms23158613. Int J Mol Sci. 2022. PMID: 35955746 Free PMC article. Review.
-
Regression of replication forks stalled by leading-strand template damage: II. Regression by RecA is inhibited by SSB.J Biol Chem. 2014 Oct 10;289(41):28388-98. doi: 10.1074/jbc.M114.587907. Epub 2014 Aug 19. J Biol Chem. 2014. PMID: 25138217 Free PMC article.
-
Characterization of the ATPase activity of RecG and RuvAB proteins on model fork structures reveals insight into stalled DNA replication fork repair.J Biol Chem. 2013 Sep 13;288(37):26397-409. doi: 10.1074/jbc.M113.500223. Epub 2013 Jul 27. J Biol Chem. 2013. PMID: 23893472 Free PMC article.
-
Formation of Holliday junctions by regression of nascent DNA in intermediates containing stalled replication forks: RecG stimulates regression even when the DNA is negatively supercoiled.Proc Natl Acad Sci U S A. 2001 Jul 17;98(15):8235-40. doi: 10.1073/pnas.121007798. Proc Natl Acad Sci U S A. 2001. PMID: 11459958 Free PMC article.
-
DNA Helicase-SSB Interactions Critical to the Regression and Restart of Stalled DNA Replication forks in Escherichia coli.Genes (Basel). 2020 Apr 26;11(5):471. doi: 10.3390/genes11050471. Genes (Basel). 2020. PMID: 32357475 Free PMC article. Review.
Cited by
-
Single-molecule insight into stalled replication fork rescue in Escherichia coli.Nucleic Acids Res. 2021 May 7;49(8):4220-4238. doi: 10.1093/nar/gkab142. Nucleic Acids Res. 2021. PMID: 33744948 Free PMC article. Review.
-
RecG controls DNA amplification at double-strand breaks and arrested replication forks.FEBS Lett. 2017 Apr;591(8):1101-1113. doi: 10.1002/1873-3468.12583. Epub 2017 Feb 28. FEBS Lett. 2017. PMID: 28155219 Free PMC article. Review.
-
Mycobacterium tuberculosis RecG protein but not RuvAB or RecA protein is efficient at remodeling the stalled replication forks: implications for multiple mechanisms of replication restart in mycobacteria.J Biol Chem. 2015 Oct 2;290(40):24119-39. doi: 10.1074/jbc.M115.671164. Epub 2015 Aug 14. J Biol Chem. 2015. PMID: 26276393 Free PMC article.
-
The Biochemical Mechanism of Fork Regression in Prokaryotes and Eukaryotes-A Single Molecule Comparison.Int J Mol Sci. 2022 Aug 3;23(15):8613. doi: 10.3390/ijms23158613. Int J Mol Sci. 2022. PMID: 35955746 Free PMC article. Review.
-
SSB and the RecG DNA helicase: an intimate association to rescue a stalled replication fork.Protein Sci. 2017 Apr;26(4):638-649. doi: 10.1002/pro.3114. Epub 2017 Mar 17. Protein Sci. 2017. PMID: 28078722 Free PMC article. Review.
References
-
- Hanawalt P. C. (1966) The U.V. sensitivity of bacteria: its relation to the DNA replication cycle. Photochem. Photobiol. 5, 1–12 - PubMed
-
- Skalka A. (1974) in Mechanisms of Recombination (Grell R. F., ed) pp. 421–432, Plenum Press, New York
-
- Zavitz K. H., Marians K. J. (1991) Dissecting the functional role of PriA protein-catalysed primosome assembly in Escherichia coli DNA replication. Mol. Microbiol. 5, 2869–2873 - PubMed
-
- Cox M. M., Goodman M. F., Kreuzer K. N., Sherratt D. J., Sandler S. J., Marians K. J. (2000) The importance of repairing stalled replication forks. Nature 404, 37–41 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

