Brain tumor cells in circulation are enriched for mesenchymal gene expression
- PMID: 25139148
- PMCID: PMC4221467
- DOI: 10.1158/2159-8290.CD-14-0471
Brain tumor cells in circulation are enriched for mesenchymal gene expression
Abstract
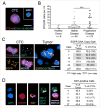
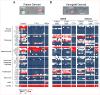
Glioblastoma (GBM) is a highly aggressive brain cancer characterized by local invasion and angiogenic recruitment, yet metastatic dissemination is extremely rare. Here, we adapted a microfluidic device to deplete hematopoietic cells from blood specimens of patients with GBM, uncovering evidence of circulating brain tumor cells (CTC). Staining and scoring criteria for GBM CTCs were first established using orthotopic patient-derived xenografts (PDX), and then applied clinically: CTCs were identified in at least one blood specimen from 13 of 33 patients (39%; 26 of 87 samples). Single GBM CTCs isolated from both patients and mouse PDX models demonstrated enrichment for mesenchymal over neural differentiation markers compared with primary GBMs. Within primary GBMs, RNA in situ hybridization identified a subpopulation of highly migratory mesenchymal tumor cells, and in a rare patient with disseminated GBM, systemic lesions were exclusively mesenchymal. Thus, a mesenchymal subset of GBM cells invades the vasculature and may proliferate outside the brain.
Significance: GBMs are locally invasive within the brain but rarely metastasize to distant organs, exemplifying the debate over "seed" versus "soil." We demonstrate that GBMs shed CTCs with invasive mesenchymal characteristics into the circulation. Rare metastatic GBM lesions are primarily mesenchymal and show additional mutations absent in the primary tumor.
©2014 American Association for Cancer Research.
Conflict of interest statement
Figures


References
-
- Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. JAMA : the journal of the American Medical Association. 2013;310:1842–50. - PubMed
-
- Smith DR, Hardman JM, Earle KM. Contiguous glioblastoma multiforme and fibrosarcoma with extracranial metastasis. Cancer. 1969;24:270–6. - PubMed
-
- Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nature reviews Cancer. 2003;3:453–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

