Molecular features and survival outcomes of the intrinsic subtypes within HER2-positive breast cancer
- PMID: 25139534
- PMCID: PMC4151853
- DOI: 10.1093/jnci/dju152
Molecular features and survival outcomes of the intrinsic subtypes within HER2-positive breast cancer
Abstract
Background: The clinical impact of the biological heterogeneity within HER2-positive (HER2+) breast cancer is not fully understood. Here, we evaluated the molecular features and survival outcomes of the intrinsic subtypes within HER2+ breast cancer.
Methods: We interrogated The Cancer Genome Atlas (n = 495) and Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) datasets (n = 1730) of primary breast cancers for molecular data derived from DNA, RNA and protein, and determined intrinsic subtype. Clinical HER2 status was defined according to American Society of Clinical Oncology (ASCO)/College of American Pathologists (CAP) guidelines or DNA copy-number aberration by single nucleotide polymorphism arrays. Cox models tested the prognostic significance of each variable in patients not treated with trastuzumab (n = 1711).
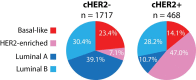
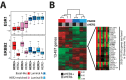
Results: Compared with clinically HER2 (cHER2)-negative breast cancer, cHER2+ breast cancer had a higher frequency of the HER2-enriched (HER2E) subtype (47.0% vs 7.1%) and a lower frequency of Luminal A (10.7% vs 39.0%) and Basal-like (14.1% vs 23.4%) subtypes. The likelihood of cHER2-positivity in HER2E, Luminal B, Basal-like and Luminal A subtypes was 64.6%, 20.0%, 14.4% and 7.3%, respectively. Within each subtype, only 0.3% to 3.9% of genes were found differentially expressed between cHER2+ and cHER2-negative tumors. Within cHER2+ tumors, HER2 gene and protein expression was statistically significantly higher in the HER2E and Basal-like subtypes than either luminal subtype. Neither cHER2 status nor the new 10-subtype copy number-based classification system (IntClust) added independent prognostic value to intrinsic subtype.
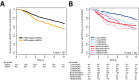
Conclusions: When the intrinsic subtypes are taken into account, cHER2-positivity does not translate into large changes in the expression of downstream signaling pathways, nor does it affect patient survival in the absence of HER2 targeting.
© The Author 2014. Published by Oxford University Press.
Figures
References
-
- Baselga J. Why the epidermal growth factor receptor? The rationale for cancer therapy. Oncologist. 2002;7(Suppl 4):2–8. - PubMed
-
- Slamon D, Clark G, Wong S, et al. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235(4785): 177–182. - PubMed
-
- Cobleigh MA, Vogel CL, Tripathy D, et al. Multinational Study of the Efficacy and Safety of Humanized Anti-HER2 Monoclonal Antibody in Women Who Have HER2-Overexpressing Metastatic Breast Cancer That Has Progressed After Chemotherapy for Metastatic Disease. J Clin Oncol. 1999;17(9):2639–2648. - PubMed
-
- Slamon DJ, Leyland-Jones B, Shak S, et al. Use of Chemotherapy plus a Monoclonal Antibody against HER2 for Metastatic Breast Cancer That Overexpresses HER2. N Engl J Med. 2001; 344(11):783–792. - PubMed
-
- Piccart-Gebhart MJ, Procter M, Leyland-Jones B, et al. Trastuzumab after Adjuvant Chemotherapy in HER2-Positive Breast Cancer. N Engl J Med. 2005;353(16):1659–1672. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

