Reconstruction and minimal gene requirements for the alternative iron-only nitrogenase in Escherichia coli
- PMID: 25139995
- PMCID: PMC4156695
- DOI: 10.1073/pnas.1411185111
Reconstruction and minimal gene requirements for the alternative iron-only nitrogenase in Escherichia coli
Abstract
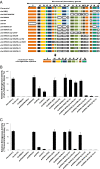
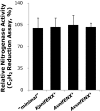
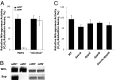
All diazotrophic organisms sequenced to date encode a molybdenum-dependent nitrogenase, but some also have alternative nitrogenases that are dependent on either vanadium (VFe) or iron only (FeFe) for activity. In Azotobacter vinelandii, expression of the three different types of nitrogenase is regulated in response to metal availability. The majority of genes required for nitrogen fixation in this organism are encoded in the nitrogen fixation (nif) gene clusters, whereas genes specific for vanadium- or iron-dependent diazotophy are encoded by the vanadium nitrogen fixation (vnf) and alternative nitrogen fixation (anf) genes, respectively. Due to the complexities of metal-dependent regulation and gene redundancy in A. vinelandii, it has been difficult to determine the precise genetic requirements for alternative nitrogen fixation. In this study, we have used Escherichia coli as a chassis to build an artificial iron-only (Anf) nitrogenase system composed of defined anf and nif genes. Using this system, we demonstrate that the pathway for biosynthesis of the iron-only cofactor (FeFe-co) is likely to be simpler than the pathway for biosynthesis of the molybdenum-dependent cofactor (FeMo-co) equivalent. A number of genes considered to be essential for nitrogen fixation by FeFe nitrogenase, including nifM, vnfEN, and anfOR, are not required for the artificial Anf system in E. coli. This finding has enabled us to engineer a minimal FeFe nitrogenase system comprising the structural anfHDGK genes and the nifBUSV genes required for metallocluster biosynthesis, with nifF and nifJ providing electron transport to the alternative nitrogenase. This minimal Anf system has potential implications for engineering diazotrophy in eukaryotes, particularly in compartments (e.g., organelles) where molybdenum may be limiting.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Xu G, Fan X, Miller AJ. Plant nitrogen assimilation and use efficiency. Annu Rev Plant Biol. 2012;63:153–182. - PubMed
-
- Oldroyd GE, Dixon R. Biotechnological solutions to the nitrogen problem. Curr Opin Biotechnol. 2014;26:19–24. - PubMed
-
- Eady RR. Structure-function relationships of alternative nitrogenases. Chem Rev. 1996;96(7):3013–3030. - PubMed
-
- Dilworth MJ, Eady RR, Eldridge ME. The alternative nitrogenase of Azotobacter chroococcum is a vanadium enzyme. Nature. 1986;322(6077):388–390.
-
- Müller A, Schneider K, Knüttel K, Hagen WR. EPR spectroscopic characterization of an ‘iron only’ nitrogenase. S = 3/2 spectrum of component 1 isolated from Rhodobacter capsulatus. FEBS Lett. 1992;303(1):36–40. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

