Flexibility in crosstalk between H2B ubiquitination and H3 methylation in vivo
- PMID: 25141862
- PMCID: PMC4253848
- DOI: 10.15252/embr.201438793
Flexibility in crosstalk between H2B ubiquitination and H3 methylation in vivo
Erratum in
- EMBO Rep. 2014 Nov;15(11):1220-1
Abstract
Histone H2B ubiquitination is a dynamic modification that promotes methylation of histone H3K79 and H3K4. This crosstalk is important for the DNA damage response and has been implicated in cancer. Here, we show that in engineered yeast strains, ubiquitins tethered to every nucleosome promote H3K79 and H3K4 methylation from a proximal as well as a more distal site, but only if in a correct orientation. This plasticity indicates that the exact location of the attachment site, the native ubiquitin-lysine linkage and ubiquitination cycles are not critical for trans-histone crosstalk in vivo. The flexibility in crosstalk also indicates that other ubiquitination events may promote H3 methylation.
Keywords: Dot1; Set1; chromatin; crosstalk; histone ubiquitination.
© 2014 The Authors.
Figures
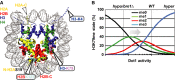
Nucleosome core particle in which H2BK123, H3K79 and H3K4 are indicated as well as the sites of linear ubiquitin attachment described below. Most of the unstructured N- and C-terminal histone tails are absent from this structure; the histone ends fused to ubiquitin (see below) are indicated with dashed triangles.
Dependence of H3K79me levels on Dot1 activity, modified from [19]. Dashed lines indicate approximate H3K79me levels in WT or bre1Δ cells. Arrow indicates the direction of the changes expected upon positive histone crosstalk.

Cartoon of H2A/H2B in the used strains.
Immunoblot analysis shows tagged H2A and the absence of endogenous wild-type H2A. Asterisk indicates non-specific signal. ‘HA indicates ubHA-H2A proteins that lost the ub moiety due to the activity of ubiquitin-specific proteases.
MS analysis of H3K79 methylation levels in strains shown in (A) and (B) (mean and individual data points of two biological replicates). See also Supplementary Fig S1F.
ChIP-qPCR data of the different methylation states at three loci. Values for each methyl antibody were normalized to H3, after which all values were normalized per methyl antibody to wild-type at the HML locus. Primer sets against HML, the promoter of the GAL1 gene and the coding sequence of SPA2 were used. Mean ± SD of three biological replicates is shown.
Immunoblot analysis of the in vitro methyltransferase activity of purified 10× His-tagged yeast or human Dot1 protein toward chromatin templates isolated from yeast strains lacking Dot1 and Bre1 and expressing wild-type H2A or N-terminal fusions of H2A. See also Supplementary Fig S1H.
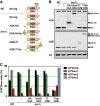
Cartoon of H2A/H2B in the used strains.
Immunoblot analysis of tagged and untagged histones. Asterisk indicates non-specific signal.
MS analysis of H3K79 methylation levels in strains expressing H2A or H2B C-terminal ub fusions (mean and individual data points of two biological replicates). See also Supplementary Fig S2C.

Immunoblot analysis of H3K4 methylation in strains expressing various histone-ubiquitin fusions.
Immunoblot analysis of activation of the central DNA damage checkpoint kinase Rad53 upon exposure of yeast cells arrested in G1/S to 100 J m−2 UV irradiation. Rad53 is activated by phosphorylation, which leads to a mobility shift.

References
-
- Zhu B, Zheng Y, Pham A-D, Mandal SS, Erdjument-Bromage H, Tempst P, Reinberg D. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol Cell. 2005;20:601–611. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

