APOBEC3F determinants of HIV-1 Vif sensitivity
- PMID: 25142588
- PMCID: PMC4248952
- DOI: 10.1128/JVI.02362-14
APOBEC3F determinants of HIV-1 Vif sensitivity
Abstract
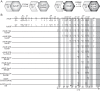
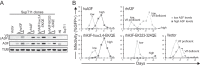
HIV-1 Vif counteracts restrictive APOBEC3 proteins by targeting them for proteasomal degradation. To determine the regions mediating sensitivity to Vif, we compared human APOBEC3F, which is HIV-1 Vif sensitive, with rhesus APOBEC3F, which is HIV-1 Vif resistant. Rhesus-human APOBEC3F chimeras and amino acid substitution mutants were tested for sensitivity to HIV-1 Vif. This approach identified the α3 and α4 helices of human APOBEC3F as important determinants of the interaction with HIV-1 Vif.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures


References
-
- Janini M, Rogers M, Birx DR, McCutchan FE. 2001. Human immunodeficiency virus type 1 DNA sequences genetically damaged by hypermutation are often abundant in patient peripheral blood mononuclear cells and may be generated during near-simultaneous infection and activation of CD4(+) T cells. J. Virol. 75:7973–7986. 10.1128/JVI.75.17.7973-7986.2001. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

