Persistence of anti-desmoglein 3 IgG(+) B-cell clones in pemphigus patients over years
- PMID: 25142730
- PMCID: PMC4294994
- DOI: 10.1038/jid.2014.291
Persistence of anti-desmoglein 3 IgG(+) B-cell clones in pemphigus patients over years
Abstract
Pemphigus vulgaris (PV) is a prototypic tissue-specific autoantibody-mediated disease, in which anti-desmoglein 3 (Dsg3) IgG autoantibodies cause life-threatening blistering. We characterized the autoimmune B-cell response over 14 patient years in two patients with active and relapsing disease, then in one of these patients after long-term remission induced by multiple courses of rituximab (anti-CD20 antibody). Characterization of the anti-Dsg3 IgG(+) repertoire by antibody phage display (APD) and PCR indicated that six clonal lines persisted in patient 1 (PV3) over 5.5 years, with only one new clone detected. Six clonal lines persisted in patient 2 (PV1) for 4 years, of which five persisted for another 4.5 years without any new clones detected. However, after long-term clinical and serologic remission, ∼11 years after initial characterization, we could no longer detect any anti-Dsg3 clones in PV1 by APD. Similarly, in another PV patient, ∼4.5 years after a course of rituximab that induced long-term remission, anti-Dsg3 B-cell clones were undetectable. These data suggest that in PV a given set of non-tolerant B-cell lineages causes autoimmune diseases and that new sets do not frequently or continually escape tolerance. Therapy such as rituximab, aimed at eliminating these aberrant sets of lineages, may be effective for disease because new ones are unlikely to develop.
Conflict of interest statement
The authors state no conflict of interest.
Figures
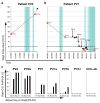

Comment in
-
Clonal analysis of B-cell response in pemphigus course: toward more effective therapies.J Invest Dermatol. 2015 Mar;135(3):651-654. doi: 10.1038/jid.2014.499. J Invest Dermatol. 2015. PMID: 25666671
References
-
- Of men, not mice. Nat Med. 2013;19:379. - PubMed
-
- Ahmed AR, Spigelman Z, Cavacini LA, et al. Treatment of pemphigus vulgaris with rituximab and intravenous immune globulin. N Engl J Med. 2006;355:1772–9. - PubMed
-
- Almugairen N, Hospital V, Bedane C, et al. Assessment of the rate of long-term complete remission off therapy in patients with pemphigus treated with different regimens including medium- and high-dose corticosteroids. J Am Acad Dermatol. 2013;69:583–8. - PubMed
-
- Barbas CF., III . Phage Display: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

