OptiType: precision HLA typing from next-generation sequencing data
- PMID: 25143287
- PMCID: PMC4441069
- DOI: 10.1093/bioinformatics/btu548
OptiType: precision HLA typing from next-generation sequencing data
Abstract
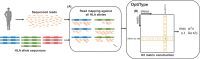
Motivation: The human leukocyte antigen (HLA) gene cluster plays a crucial role in adaptive immunity and is thus relevant in many biomedical applications. While next-generation sequencing data are often available for a patient, deducing the HLA genotype is difficult because of substantial sequence similarity within the cluster and exceptionally high variability of the loci. Established approaches, therefore, rely on specific HLA enrichment and sequencing techniques, coming at an additional cost and extra turnaround time.
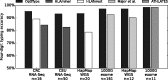
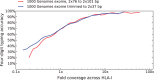
Result: We present OptiType, a novel HLA genotyping algorithm based on integer linear programming, capable of producing accurate predictions from NGS data not specifically enriched for the HLA cluster. We also present a comprehensive benchmark dataset consisting of RNA, exome and whole-genome sequencing data. OptiType significantly outperformed previously published in silico approaches with an overall accuracy of 97% enabling its use in a broad range of applications.
© The Author 2014. Published by Oxford University Press.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

