Stress-dependent proteolytic processing of the actin assembly protein Lsb1 modulates a yeast prion
- PMID: 25143386
- PMCID: PMC4183801
- DOI: 10.1074/jbc.M114.582429
Stress-dependent proteolytic processing of the actin assembly protein Lsb1 modulates a yeast prion
Abstract
Yeast prions are self-propagating amyloid-like aggregates of Q/N-rich protein that confer heritable traits and provide a model of mammalian amyloidoses. [PSI(+)] is a prion isoform of the translation termination factor Sup35. Propagation of [PSI(+)] during cell division under normal conditions and during the recovery from damaging environmental stress depends on cellular chaperones and is influenced by ubiquitin proteolysis and the actin cytoskeleton. The paralogous yeast proteins Lsb1 and Lsb2 bind the actin assembly protein Las17 (a yeast homolog of human Wiskott-Aldrich syndrome protein) and participate in the endocytic pathway. Lsb2 was shown to modulate maintenance of [PSI(+)] during and after heat shock. Here, we demonstrate that Lsb1 also regulates maintenance of the Sup35 prion during and after heat shock. These data point to the involvement of Lsb proteins in the partitioning of protein aggregates in stressed cells. Lsb1 abundance and cycling between actin patches, endoplasmic reticulum, and cytosol is regulated by the Guided Entry of Tail-anchored proteins pathway and Rsp5-dependent ubiquitination. Heat shock-induced proteolytic processing of Lsb1 is crucial for prion maintenance during stress. Our findings identify Lsb1 as another component of a tightly regulated pathway controlling protein aggregation in changing environments.
Keywords: Actin; Endoplasmic Reticulum (ER); Heat Shock; Prion; Proteasome; Rsp5; Sup35; Ubiquitylation (Ubiquitination).
© 2014 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
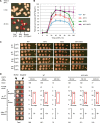

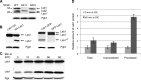

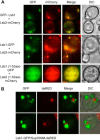


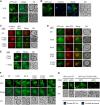
References
-
- Kopito R. R. (2000) Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 10, 524–530 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

