Synergies between Aip1p and capping protein subunits (Acp1p and Acp2p) in clathrin-mediated endocytosis and cell polarization in fission yeast
- PMID: 25143407
- PMCID: PMC4230613
- DOI: 10.1091/mbc.E13-01-0005
Synergies between Aip1p and capping protein subunits (Acp1p and Acp2p) in clathrin-mediated endocytosis and cell polarization in fission yeast
Abstract
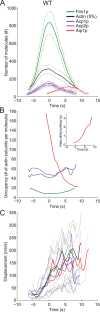
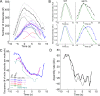
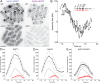
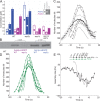
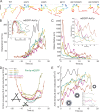
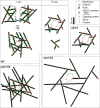
Aip1p cooperates with actin-depolymerizing factor (ADF)/cofilin to disassemble actin filaments in vitro and in vivo, and is proposed to cap actin filament barbed ends. We address the synergies between Aip1p and the capping protein heterodimer Acp1p/Acp2p during clathrin-mediated endocytosis in fission yeast. Using quantitative microscopy and new methods we have developed for data alignment and analysis, we show that heterodimeric capping protein can replace Aip1p, but Aip1p cannot replace capping protein in endocytic patches. Our quantitative analysis reveals that the actin meshwork is organized radially and is compacted by the cross-linker fimbrin before the endocytic vesicle is released from the plasma membrane. Capping protein and Aip1p help maintain the high density of actin filaments in meshwork by keeping actin filaments close enough for cross-linking. Our experiments also reveal new cellular functions for Acp1p and Acp2p independent of their capping activity. We identified two independent pathways that control polarization of endocytic sites, one depending on acp2(+) and aip1(+) during interphase and the other independent of acp1(+), acp2(+), and aip1(+) during mitosis.
© 2014 Berro and Pollard. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures

References
-
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, III, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. - PubMed
-
- Basi G, Schmid E, Maundrell K. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene. 1993;123:131–136. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

