Activation of TRPML1 clears intraneuronal Aβ in preclinical models of HIV infection
- PMID: 25143627
- PMCID: PMC4138351
- DOI: 10.1523/JNEUROSCI.0210-14.2014
Activation of TRPML1 clears intraneuronal Aβ in preclinical models of HIV infection
Abstract
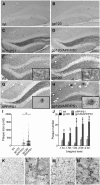
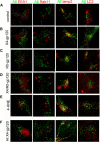
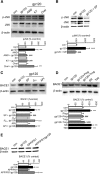
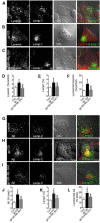
Antiretroviral therapy extends the lifespan of human immunodeficiency virus (HIV)-infected patients, but many survivors develop premature impairments in cognition. These residual cognitive impairments may involve aberrant deposition of amyloid β-peptides (Aβ). By unknown mechanisms, Aβ accumulates in the lysosomal and autophagic compartments of neurons in the HIV-infected brain. Here we identify the molecular events evoked by the HIV coat protein gp120 that facilitate the intraneuronal accumulation of Aβ. We created a triple transgenic gp120/APP/PS1 mouse that recapitulates intraneuronal deposition of Aβ in a manner reminiscent of the HIV-infected brain. In cultured neurons, we found that the HIV coat protein gp120 increased the transcriptional expression of BACE1 through repression of PPARγ, and increased APP expression by promoting interaction of the translation-activating RBP heterogeneous nuclear ribonucleoprotein C with APP mRNA. APP and BACE1 were colocalized into stabilized membrane microdomains, where the β-cleavage of APP and Aβ formation were enhanced. Aβ-peptides became localized to lysosomes that were engorged with sphingomyelin and calcium. Stimulating calcium efflux from lysosomes with a TRPM1 agonist promoted calcium efflux, luminal acidification, and cleared both sphingomyelin and Aβ from lysosomes. These findings suggest that therapeutics targeted to reduce lysosomal pH in neurodegenerative conditions may protect neurons by facilitating the clearance of accumulated sphingolipids and Aβ-peptides.
Keywords: HIV; amyloid; dementia; endosome; lysosome; neuron.
Copyright © 2014 the authors 0270-6474/14/3411485-19$15.00/0.
Figures








References
-
- Akay C, Lindl KA, Shyam N, Nabet B, Goenaga-Vazquez Y, Ruzbarsky J, Wang Y, Kolson DL, Jordan-Sciutto KL. Activation status of integrated stress response pathways in neurones and astrocytes of HIV-associated neurocognitive disorders (HAND) cortex. Neuropathol Appl Neurobiol. 2012;38:175–200. doi: 10.1111/j.1365-2990.2011.01215.x. - DOI - PMC - PubMed
-
- Ances BM, Christensen JJ, Teshome M, Taylor J, Xiong C, Aldea P, Fagan AM, Holtzman DM, Morris JC, Mintun MA, Clifford DB. Cognitively unimpaired HIV-positive subjects do not have increased 11C-PiB: a case-control study. Neurology. 2010;75:111–115. doi: 10.1212/WNL.0b013e3181e7b66e. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AA0017408/AA/NIAAA NIH HHS/United States
- P30 MH075673/MH/NIMH NIH HHS/United States
- AG043338/AG/NIA NIH HHS/United States
- R21 AG034849/AG/NIA NIH HHS/United States
- GM103329/GM/NIGMS NIH HHS/United States
- R01 NS062792/NS/NINDS NIH HHS/United States
- R01 MH077542/MH/NIMH NIH HHS/United States
- AG034849/AG/NIA NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- MH075673/MH/NIMH NIH HHS/United States
- R21 AG043338/AG/NIA NIH HHS/United States
- MH077542/MH/NIMH NIH HHS/United States
- R01 AA017408/AA/NIAAA NIH HHS/United States
- R01 MH096636/MH/NIMH NIH HHS/United States
- R01 MH100972/MH/NIMH NIH HHS/United States
- P30GM103329/GM/NIGMS NIH HHS/United States
- P30 GM103329/GM/NIGMS NIH HHS/United States
- R01MH100972/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
